I had a pretty rough month and a shortage of both time and enthusiasm. As I was thinking about not getting up at 4am to have a couple hours to do cool science, I saw this paper in my Scholar Alerts.
About f'ing time! I honestly thought I was going to have to buy a hybrid to do this one myself.
Ions pass BY the Orbitrap detector. They do not strike the Orbitrap detector. As such, they are always at a sensitivity disadvantage to virtually anything where ions make contact with the detector.
Hey! I learned how to make a GIF!You can say a lot of bad things about ion traps. Bad mass accuracy. Low resolution. The spectra from newer ion traps look terrible compared to the ones of the past. Et cetera. But they are sensitive!
And if you are into the MS3 based quantification methods for TMT/iTRAQ quantification, we've known for a long time that the ion trap can be a serious asset.
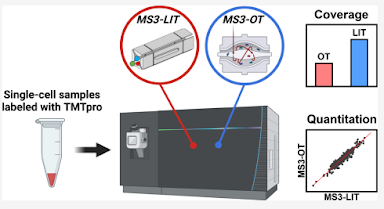
This group broke out the CellenOne and did the NanoPots thing, prepped some real human cells and tried it out. The results are predictably impressive. Higher sequence coverage and more overall proteins identified than with the Orbitrap based quan. However, the quantification distinction at a protein level didn't appear to be as good. (Some differential proteins don't appear significant when using the ion trap). At a functional level the results line up better between the different methods, but there are downsides.
Worth considering, they used a relatively small sample set. I think they did around 100 cells by the ion trap MS3 method. You'd have to wonder if you'd see the same weaknesses at 1,000 cells.
This is a great study that I was hoping someone would do for a while so we could see what it looked like and it is really awesome to see this group execute it to perfection.
If you've got an old hybrid sitting around and are on the fence about getting rid of it, you should 100% check this out. I don't have data to support this, but I strongly suspect that the older LTQ Orbitraps with the ion trap on the front of the instrument would be amazing tools for this application. Sure, you can't SPS on them (without getting some magic software from some guys in Boston or altering the software) but you can isolate the most abundant high mass fragment ion from the MS2 and that should work well.
A really cool thing about single cell proteomics has been how it has sort of flipped the paradigm on instrument selections. Virtually all of the data we've seen from Slavov lab has been on the plain old Q Exactive Classic that data has been right there with literally any other data we've seen from instruments that cost 4x as much. This study opens up SCP to an entire generation of instruments.

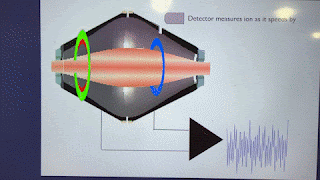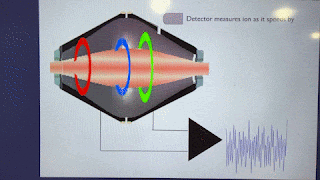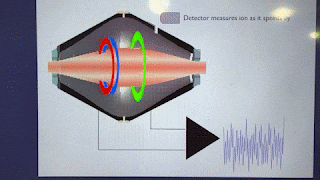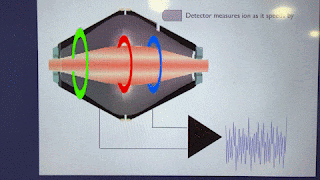
Love to see some ion trap good news. However, this statement is false: "The spectra from newer ion traps look terrible compared to the ones of the past". Fusion had the same traps as Velos which had better resolution than LTQ, but got a faster electrometer, which allowed to resolve the 125 kDa/s peaks a little better. Newer generations of Tribrid got bigger trap chamber turbos, lowering the base pressure and improving resolution further. Ion statistics noise got lower with each generation of brighter source, improving spectral quality. Look at the peaks on a Tribrid while it calibrates, they're really skinny for the amount of time spent analyzing each peak...unit resolution with < 8 microseconds analysis time per peak? It's amazing.
ReplyDelete