Saturday, February 25, 2023
Drag that LTQ out of your garage -- single cell proteomics time!
Thursday, February 23, 2023
Clinical knowledge graph! Open notebooks to help us take that next step!
What's a Jupyter notebook? It's like a really elaborate programming exercise with all the notes so you can go through and follow the steps 1 at a time. If they're done right even I can follow them. If they are done wrong....well....I can't rule out my own inabilities, but it's awesome when they're right. You feel really smart at the end and like buying whoever wrote it a beer sometime.
Tuesday, February 21, 2023
DirectLFQ - Super fast requantification of your processed data for some reason!
I'm more than a little confused about this, but I tell you what -- it is FAST!
I read the preprint sort of asleep early this morning, then this guy drove by my house with CRAZY LOUD BASS, like you could feel it in your chest sort of bass when I was bringing my trashcans in. I waved him down and told him I also love the Backstreet Boys and for some reason it made him really really mad. Apparently Backstreet is NOT back and I misidentified the artist. Surprised he could tell what he was playing, but that's okay. I was a little more alert after this discussion got really heated in a hurry, so I looked back through the paper.
I downloaded it (you can get the one-click GUI's here) and I ran it.
I pulled up 128 different mouse organs that I'd ran on a TIMSTOF in DDA format and processed with FragPipe 19.1 last week. I loaded the combined_ion.tsv file into DirectDIA and within about 60- 90 seconds it made me a new protein report. I didn't even have time to get distracted.
Where I started to get confused is here -- I already have a protein report from FragPipe. When I compare them, they are pretty similar. DirectLFQ had about 10% more missing values at the protein level (low n) but the numbers do seem to line up reasonably well.
My big question is where this fits into my workflow? I don't really have tools here where I get PSM level quan and I don't get protein level quan. The paper doesn't illuminate it for me much. I do get it, the software definitely is fast, I just can't figure out when I'd use it?
You can load DIA-NN data into it and MaxQuant data and there is a generic format uploader for whatever else you want to requantify. DIA-NN and MaxQuant both also make protein level quan reports.
If I look at my Fragpipe log files the assembly of the PSM level quan data to protein level isn't exactly my limiting step either:
It looks like this took about 2 minutes?
Look, I'm not taking anything away from this. It is free software that runs on Linux, Mac, PC and it can take your PSM level quan data and make you a protein quan report. If you don't have that capability in your pipeline -- you do now!
Monday, February 20, 2023
You asked for it! Install and run Proteome Discoverer on a Mac!
What's that pointing at? Weird symbol.
Sunday, February 19, 2023
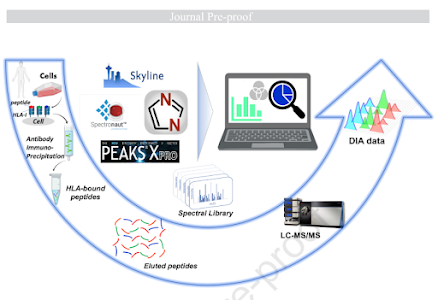
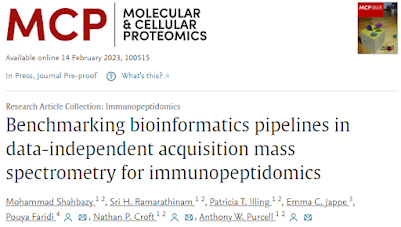
Benchmarking pipelines for DIA immunopeptidomics!
Immunopeptidomics is somehow even tougher to do than to spell.
The basic residues (if they are there at all) typically are not at a termini.
You may see a lot of peptides that only go to z = 1.
They often fragment like crap.
Should you go ahead and do it with DIA? (Yikes. Y'all try it first and write up how it went and I'll think about it). Well...here you go!
PEAKS vs Skyline vs DIA-NN vs SpectroNaut!
Who comes out on top? To be honest, it is sort of a mixed bag with pros and cons of each.
Figure 3 is a highlight for me because the consistency between all 4 is a whole lot higher than I would have guessed. The numbers are all within about 10% of each other.
Saturday, February 18, 2023
Learn about protein 3D structures in 3D cell cultures with spheroid FPOP!
Imma just quote the abstract:
Many cancer drugs fail at treating solid epithelial tumors with hypoxia and insufficient drug penetration thought to be contributing factors to the observed chemoresistance. Owing to this, it is imperative to evaluate potential cancer drugs in conditions as close to in vivo as possible, which is not always done
We've known for years and years and years that cells growing in 2D on a plate have limitations in terms of representation actual biology.
3D cultures also have limitations (and so do animal models and so do human studies) so we do the best we can.
One of the limitations in any 3D culture study I've ever seen is super low N
SO. What if you use Hummon lab's procedures for creating high numbers of tiny 3D cultures for proteomics for cancer drugs AND Lisa Jones lab's approaches for in cell photo-oxidation of proteins and working out 3D cultures?
Could you get an understanding of the GLOBAL PROTEIN CONFORMATIONS OF 3D CELL CULTURES TREATED WITH DRUGS en masse?
Hell yes you could. Check this out!
They get structural information by FPOP on as many as 635 proteins at a time! They also throw in some pretty MALDI imaging stuff to show how far the FPOP radicals and stuff get into the cells!
Ridiculously great study from beginning to end.
HeadbangForScience.org
Need an organization to get behind in 2023? Check out this killer scholarship founded by the metal ambassador himself, Jose Mangin. They line up some serious acts for free shows to raise money for the scholarship fund. Given the prevalence of metal fans in proteomics, there is a decent chance this scholarship would strengthen our field down the road.
Friday, February 17, 2023
Today's Proteomics Show Road to HUPO features Dr. Alexander Ivanov!
This week's episode just launched, and there is a funny story behind it. Neely was in his car for the podcast during the arctic blast thing near the holidays. Dude was wearing a shirt with long sleeves because Charleston was a frigid 54F (12.2C).
Alexander Ivanov (lab website here) was great while we did our level best to screw up his podcast.
As an aside, I have google scholar alerts on some people in our field. Maybe a lot of people. The good ones, at least. And this was really funny to me, because I totally thought they got this name wrong --
Wrong one! Anyway, this is me rambling in the orange box so I felt like something I did today worked!
Thursday, February 16, 2023
You can now directly transfer bioRxiv to JPR!
For a while now there has been just one major publisher that can't diretly take biorXiv transfers.
NO MORE! As of today you can upload your preprint to biorXIV and then seamlessly (seemlessly? semleesly?) transfer that over to those nitpicky nerds at JPR!
Monday, February 13, 2023
Real time search with a twist -- predicted and empirical spectral libraries!
I SHOULD NOT BE BLOGGING. I SHOULD BE DOING GRANT PAPERWORK.
I CAN NOT READ THIS. SCHOLAR, QUIT SENDING ME COOL STUFF.
I WILL LATER.
Sunday, February 12, 2023
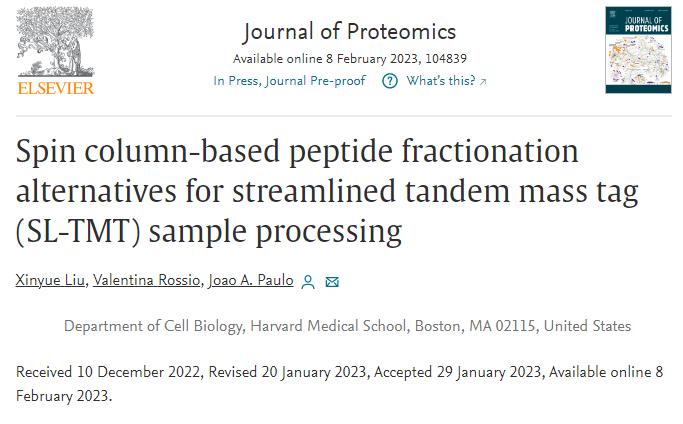
High pH fractionation isn't cutting it? There are easy strong anion exchange kits!
Did you know there were spin columns for strong anion exchange chromatography? I remember hearing about them 30 years ago or something, but they are still out there and they appear to work!
Sometimes we get samples in and HPH fractionation just seems to waste a lot of our time over just doing a single nonfractionated injection. I don't know why and(Rumor is there is a big ad campaign at today's StupidBowl that reunites some of this amazing cast. That's almost enough to make me keep it on in the background).
Maybe I should just get some of these SAX columns and keep them around in case I need extra coverage!
(Best I could do, everything that I have to do in 2023 has a deadline of February 15th, I should do some of it, or sleep, probably both!)
Oh yeah, they appear to have used these things -- https://www.thermofisher.com/order/catalog/product/90010
Saturday, February 11, 2023
Stop the guesswork in your DIA windows! DO-MS has been extended to (Orbitrap) DIA!
This is a little late for us for something we're writing up. I'm not redoing 700 injections or whatever with better DIA windows. I made a pool, tried some methods, processed the data, picked the two best methods and made a method that was a decent compromise between the two. I then ran three replicates with that, processed the data and it looked okay so I used that for the month or two or whatever.
Sound familiar?
Maybe DON'T DO THAT and DO-MS! Which can now do DIA (for Orbitraps, which was actually what I needed it for. The new pasefDIA methods in TIMSControl 4.0 are ridiculously good).
Or get DO-MS here.Friday, February 10, 2023
SEDA -- Turn that crappy genome or protein FASTA into something great, no coding required!
I've got a couple of projects that have been sitting around because the genomics input needs cleaned up and made into a nice concise FASTA and that stuff is
Almost all of the things require you to concisely type the exact letters into boxes where there are no pictures. As you might have noticed, I get my letters mixed up when I type and that's exactly what you shouldn't do at consoles and command lines.
What if you could just click around a really simple GUI and DO ALL THE THINGS?
Hi, SEDA, (SEquence DAtabase builder) nice to meet you!
While you should read the paper, this software is super intuitive and really easy to use and you can get it for Windows or Penguins or whatever all here.
If you've got a huge FASTA file you'll either want to turn off the in memory indexing or figure out where it hides the Java limits for allowable RAM usage. I found instructions on where to find that text file for Penguin operating systems, but I can't hunt it down for Windows. I just turned it off for a 2GB file and let it do the work off the solid state drive. I don't know how long it took, I went to look for mass spec that was last seen leaving Memphis on Tuesday. It is amazing how often 1,400 pound (635 kg) things seem to end up in the wrong states. You'd think it would be more efficient to take it directly to the correct state, but I'm clearly no cartographer.
Best parts here, you can easily 6-frame translate and remove all your redundancy all within this program. Do you know that your organism absolutely definitely doesn't use some specific start codons? You can toss those out before you even get started. These are all things you can totally do in a console or at a command line, but I've never seen in a simple and easy to install GUI.
Thursday, February 9, 2023
Huge upgrades for MetaMorpheus! Spectral library searches, library generation and top down!
Wednesday, February 8, 2023
DIA vs TMT for plasma proteomics -- a real breakdown from a real science blog!
Have some questions about how to do plasma proteomics with leading approaches?
Check out this real science blog post on Phil's Github!
If that doesn't answer your questions and you don't mind -75F weather, we'll sort the rest out in Chicago at the US HUPO LFQ PROTEOMICS BATTLE ROYALE!
For people who have asked if the latter will be recorded, I don't think we can -- for a lot of reasons I can't disclose here. It's a down to earth no holds barred breakdown of leading LFQ proteomics technologies.
Monday, February 6, 2023
Finally! A complete kit for offline fractionation in plates!
Disclaimers in addition to the ones over there somewhere --->
1) I haven't tried these yet, but I'm adding them to our ordering sheet today.
2) I don't know these people, I don't think.
AND they come in 96-well plate formats. If I can get them in house (always sketchy here, I still can't order IonOpticks columns or pay my ACS open access fees since neither are approved vendors at my university), you bet I'll be showing data from them.
Saturday, February 4, 2023
Another US HUPO 2023 guest on the podcast! This week Jenny van Eyk!
The next episode of The Road to Chicago podcast series just posted, featuring the inspirational Dr. Jenny Van Eyk.
If you were on the fence about sunny Chicago in about 30 days, let Jenny tell you not only about her work, but about the speakers and projects in real translational clinical proteomics that you'll get to see and hear about in Chicago.
Thursday, February 2, 2023
What the developers don't want you to know about MSstatsShiny! (Download an annotation template here!)
In advance I want to apologize if this post comes off soundy a little sarcastic. That isn't my intention. We all know that sometimes people who do great science are not good at communicating said science. One example might be the MSStats thing that most mass spectrometrists have heard of but is only accessible to people that commonly compile code.
MSstatsShiny has a pretty web interface but you can only use it if you have a small input file and only if you know the absolutely top secret - you can spend 3 days looking for it and you won't find it anywhere - annotation file format!
If you want to run it locally, all the steps are at the bottom of this post and you still need that annotation file format.
I broke out the big guns here and had one of the best real bioinformaticians on the planet wade through pages and pages of stuff that looks like this
Want it? Too bad, I didn't spend 3 days looking for it AND cash in a huge favor just to give it away for free.
J/K, but y'all owe me. Download it here.
With that out of the way, let's go through THE MASS SPECTROMETRISTS GUIDE TO INSTALLING AND RUNNING MSstatsShiny!
First you're going to need a pain in the butt called RStudio, and you'll need the newest one. You can get it here.
Once you install it, you'll want to open it and go to the console tab.
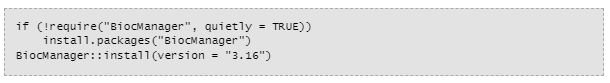
Now you're going to need bioconductor. You can get that here: https://www.bioconductor.org/install/
Bioconductor is a big package that is necessary for installing all sorts of other things. It's like how you have to install Steam to get games from the Steam store.
Anytime you see something in a grey box on Bioconductor like this, you can just copy and paste this thing and then you put it in the R console by the "greater than" sign. In this context it is probably called a carrot.
Protip for R. If your console has been doing stuff and you don't see a greater than sign, go to the top and hit the little stop sign. Chances are it is running something in the background.
Copy and paste the thing in the grey box to the lowest greater than sign on your screen and hit enter.
Now, I don't know why, but when you are installing stuff IT WILL LOOK LIKE YOU JUST TRIGGERED A NUCLEAR WAR FROM YOUR COMPUTER. You'll see what I mean. The angriest red scrolling text that you've ever seen, generally means things are going okay. Some nerd in Australia found it funny and no one has ever changed it.
Protip for R #2: What I've found with the catastrophe that Windows has become is that my program files will sometimes be locked as read only. You may have to follow the directions in the R console to the file it can't write to then right click, get the little box to pop up, go to properties and unclick "read only".
Once the stress of what a terrible thing you've obviously done is over. It's time to do it again!
Go get MSStatsShiny here! https://bioconductor.org/packages/release/bioc/html/MSstatsShiny.html
Copy the stuff in the grey box to your greater than sign and avert your eyes from the terror on your screen!
It might ask you questions about things you don't know about while you're installing. Just say yes. It will probably be fine.
NOW IF YOU GET THROUGH THIS HERE IS THE NEXT SECRET.
To run the program you'll need this top secret code: (I read somewhere that I might not need all these words, but this is what works for me)
Wednesday, February 1, 2023
3D print an LCMS tool kit for those Thermo tools you lose all the time!
How freaking great is this?!?
Do you know where all of those things are right this second? I can definitely find them all...eventually...