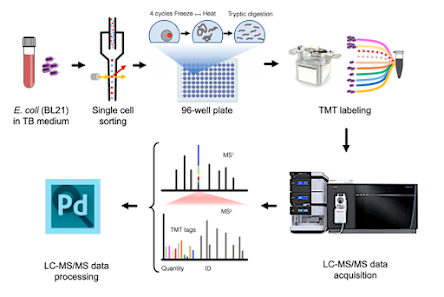
It was 100F in Vienna and the great people at the IMP put on one of the greatest meetings I've ever been to. I can't do it justice, because this meeting was just too good. How good? There were ice cream breaks and while we're still screwing around with "hey! I can see proteins in a single cell" on this side of the Atlantic, in Europe they're actually applying it to help people. For real. IMP is working with real surgeons. Pharma people were everywhere at hte meeting as well. Application, application, application!
Right before I got there the IMP received a giant check to build up their capabilities! Ever seen a giant check for science? I sure haven't!
Also -- ESCP sold out early at 200 people. It was packed.
Quick run down of day 1:
Karl Mechtler himself did a stage appearance and said a few words before handing it over to EuPA's leadership (currently in changing of the guard status) and it kicked off.
I've got to be really careful with photos and what I talk about - there was A LOT of unpublished data. I got all inspired and stayed up most of the night to change my slides and show what I was currently working on. Which...now I've definitely got to write up...but it was that sort of vibe.
If I miss anyone, it isn't a slight, I just have a real job and this isn't it.
Bogdan Budnik showed crazy new data on human neurons and drugs. He also demonstrated some really really cutting edge software his team has developed and have in works. He also very succintly walked us through the streamlined workflow his group at Harvard/Wyss uses to complete around 400 cells/day using a Q Exactive (I'm not sure which type) and an Exploris 240. Stunning data reinforcing that single cell proteomics isn't all about the hardware. Extreme paper watch alert on this one.
Lennart Martens gave basically a what's what on CompOmics -- tools I knew about and ones I didn't. If you haven't been here in a while, it's a good time to visit it. https://www.compomics.com/
James Fulcher walked us through Nano Droplet Splitting. Which is, btw, the thing that gets all the fancy genomics people excited about single cell proteomics. When I say - well....you could still get your largely useless RNA out of each of these cells and I'll still have material to do the stuff that is phenotypically relevant, they get excited. As long as they get their largely useless but exceptionally pretty pictures out of a population of cells, you've got an ally. (Wow. That sounded sorta mean. I like pretty pictures as much as anyone.)
(Fulcher, 99% sure everything on this slide is covered in his preprint)
Alexander Ivanov showed single cell top down. A lot of that has been featured on the blog and on the podcast, but I invite you to check it out.
ESCP promoted a lot of really exceptional students. Abel Vertesy, Anuar Makhmut and Suniya Khatun all showed currently unpublished data that I don't want to discuss until I see it preprinted. All reinforced how much better the next generation of scientists may be at proteomics than us.
I worked on my slides at lunch and ran over a little and missed Anjali Seth and part of Ryan Kelly's talk. We have spoke at a lot of things together over the last year or two and I feel like I'm mostly up to date.
Valdemaras Petrosius showed off the Schoof lab's fancy single cell statistics and some comparisons between Orbitrap Eclipse and Astral. Most of that has been featured on the blog somewhere. I got to speak to him after and he's been doing mass spectrometry for about 2 years or something. Scary good data 2 years in? Didn't ABRF conclude that it took about 5 for us to generate solid data? Curve appears to be steeper some places than others.
Day 1 wrapped up with Peter Pichler giving a history of surprising history of medicine/surgery with dates so far back in time we only have archaeological evidence. Then we did a cookout on the roof of the IMP!
Day 2! (Sabrina Richter on why we need proteomics when we've got single cell RNASeq and all their sophisticated informatics)
Day 2 started strong with Chris Rose (Genentech) and then Erwin Schoof who I finally got to meet in person and one of the great student talks I mentioned above and Matzinger Manuel (who has the most engaging illustration style for figures I might have ever seen -- I haven't been able to catch up with him to see if he literally draws them or if it is some AI graphics package). All of them have had work featured on this silly blog recently, and maybe I'll put in some links if I get a chance. Off to preschool in a second.
(Found a pic. How cool and engaging is that?)
Vegvari Akos (also finally got to meet, terrible taste in music, but really cool guy) went after a question that I think about a lot. Could we do less sophisticated systems? Like -- since bacteria have proteomes mabye 1% as complex as ours, could you gain enough in your reduced "noise" level to quantify things? Again, a lot unpublished, but I think it is fair to say that the number of IDs wouldn't wow anyone, but there is potential there.
Sabrina Richter (pictured above) demonstrated some super sophisticated statistics applied to single cell proteomics. That lab is just killing it.
We had a minor schedule alteration where we upgraded from Matthias Mann to Dr. Florian Rosenberger for deep visual proteomics. As always, intimidating stuff out of the Mann lab. I was a little disappointed by the resolution of the first deep visual proteomics study but the resolution they have now is absurd. Ugh. Like I really want ot type a word here and I don't think it's preprinted yet so I shouldn't. It has 4 syllables, but I can't type it. Crazy data coming.
Putting here for proof people invite me to give lectures (having some problems getting my expense reports approved).
My grandmother didn't make it through the pandemic and I miss her dearly, but I'm glad on some level she didn't live long enough to see this picture of me being seeing in public wearing 15 year old jeans. And...to be honest...maybe I need to go shopping for clothes more than once every decade....might need to work that in somewhere.
Then the conference got CRAZY. Not because of the ice cream break. Francis Berger's talk was amazing and I don't know how much I can talk about it. I've never wanted to see a live video of a brain tumor being removed. I've never wondered how many cells you can take out of a certain area in a human brain without killing someone, but it isn't many. And I've never thought about a zombie cell.
The day wrapped up with Michale Caldwell (NorthWestern) talking about the work some Kelleher guy showed at SCP in Boston I mentioned recently and Christoph Krisp (Bruker) showing what you can do at 300Hz with single cell loads (300 f'ing Hz!) and a really cool breakdown from a Nature Editor about their new directions. They're being super supportive of next generation proteomics and it was cool to hear more about their directions.
The conference wrapped up for me with dinner with some really cool new friends (and Jarrod who is the least sketchy Australian you'll ever run into).
There was a great tour of Vienna the next morning, but I was on the way to the airport so I only got to yell funny sounds from a taxi on my way out.
Okay, so maybe I'll fill in some links later to different papers, but if you took anything from this rambling today it should be that EXTREME PAPER WATCHES SHOULD BE IN EFFECT. For real, I've said before that single cell proteomics is mass spectrometries moonshot. I'll go one further
Single cell proteomics is the Moonshot project for biology and medical research. What is going to come out of it as it continues to become less "cool trick" and more application is going to change things. The bigger impact may still be what trickles down from it as people start thinking "10 cells worth of material? That's a ton!"