Sunday, February 27, 2022
The structural context of PTMs at a proteome scale....
Friday, February 25, 2022
Some ideas to maybe feel a little less useless about the unfolding war.
If you're lucky enough to just feel powerless and appalled while watching the actions of some tiny dictator, rather than scared for your homeland and your family and your life, maybe you aren't entirely powerless? I found some of this really helpful for my own personal sanity the last 36 hour or so.
World Central Kitchen is the shit. They're on the ground in Poland right now (where the borders are open and you don't need paperwork to get through!) trying to help. With operating expenses of less than 16%, almost 85% of every donation goes right to the front lines. You can pitch in here.
I'd suggest we all stop and help the Anonymous effort, but...we aren't exactly a field packed with Lis Salander level talents (I generalize, a couple of y'all are good!) but maybe right now is a great time to send takeout and caffeine to programmers that you suspect have Guy Fawkes masks hidden behind their Lego collections?
Look, I have to drop some awful attempt at comedy in here and I'm just trying to feel a little less useless about the insanity that keeps popping up on my screens.
TMTPro18-Plex method for Proteome Discoverer and exact masses!
Got your TMT18-plex reagents in house?
Need the methods? For ProteomeDiscoverer you can download this .XML here.
If you are using PD 2.4 or older you might need to update your modifications folder from UniMod. In PD 2.1 and 2.2 which I keep because some cool free tools work in those and haven't been updated to the new ones, I haven't been able to directly import from Unimod. Instead I copy the XML and then import that.
Need the exact masses for some other tool? I put them into Excel here.
126.127726
127.124761
127.131081
128.128116
128.134436
129.131471
129.13779
130.134825
130.141145
131.13818
131.1445
132.141535
132.147855
133.14489
133.15121
134.148245
134.154565
135.1516
When 134C/135N comes in I'll probably need to update MaxQuant, but I haven't done it yet.
Thursday, February 24, 2022
multiFLEX-LF -- Overcome protein/PTM stoichiometry!
Update: Paper link to the software is wrong. You want this, I've contacted the authors. one: https://github.com/SteenOmicsLab/multiFLEX-LF/releases
Wednesday, February 23, 2022
Tutorial -- highly accurate and sensitive multiplex quan on TIMSTOFs!
The last one was well received! How 'bout one that took us months to put together?
For more details, check this out:
1) You need to have a low mass fragmentation scan and a high mass fragmentation scan. This is due to TOF effects through the quad prior to pulse activation that sends the fragments into the TOF. Oddly, enough someone solved this problem recently, but I don't think they'll share it with this vendor.
End result is that your (theoretical) 120Hz instrument is going to be a 60Hz instrument. The cool part is that since you have two separate packets of fragments you can do different things with them. The obvious idea is to use a relatively low energy on the packet you'll use for sequencing your peptide and get stupid with the fragmentation energy you'll use for the packet for reporter ion release.
2) While you can do TMT18-plex quan if you want (see supplemental of preprint) it might not be completely ready yet. Remember how when the TMT10plex came out and all of us with OrbiXL/Velos systems used 35,000 MS2 (4 scans/second or something? 3.5?) and it worked okay, but anyone with the Elite system made fun of us because they got baseline separation at 60k, but no one stopped to check to see if there was any biologically relevant difference because we didn't know how but we didn't want to be made fun of by labs with vendor deals or infinite research funding so we basically stopped? Same principle here. A little worse because you can't centroid the TIMSTOF data perfectly yet. At least 2 groups are working on it. I'm eagerly awaiting final products.
I recommend using the TMTPro18 reagent and then using every other tag. We use the n tags for consistency and keep the c tags for making spectral libraries and other one-off type experiments. With the TMTPro18 you can 10-plex this way! That's the best you could do with anything just a couple of years ago!
3) The mass accuracy isn't going to be as good. OR IS IT? I'm super proud of this idea because I don't think anyone thought of it first somehow and I was like "I'll find out later someone did this in 2005 and I should have known" but -- check this out. In a TMT quan experiment you always have a reporter ion. I'm doing SCoPE-MS so my carrier channel is always this high abundance flag in every spectrum. I know the exact mass of that! Why don't I just recalibrate each file on that fragment ion? Taadaa!
I do also recommend doing the main mass calibration with a syringe using the Agilent low concentration TOF mix because it has low mass ions, prior to starting any TMT/iTRAQ batch, because if you calibrate off of the filter with your lowest mass being 622, you can get wobbly in the low mass range. This can still adjust it, but it's better to start close.
4) Adjust your TIMS to match your eluting ions and keep in mind that TMT6/10/11 and TMTPro migrate differently to each other and unlabeled peptides. If you want the best coisolation interference, use a really narrow range. 0.8-1.3 can work really well. However, if coisolation isn't the biggest concern for your sample (like there is very little around that could coisolate in your sample, crank that number up).
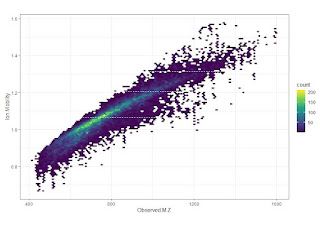
TMT11plex shown, this didn't make the paper, butI love these hexagonal R plots! Almost all of the ions are between 0.8 and 1.4, literally ever identified peptide is between 0.7 and 1.5, so even narrowing it down a little will increase you ion mobility resolution for each MS1 and MS2 spectrum.
5) Note on quad isolation -- the vendor default TMT template (which I am lobbying to be renamed pasefRiQ because that's just cooler and less concerns of lawsuits!) uses a 2Da low mass and 3 Da high mass cutoff. That works for signal intensity (you didn't buy this thing because of it's great quad...I hope....?). Using the TMT TKO standard 1.5 up/1.5 down seems to be the best in my hands, but I didn't try 1.2 or 1.26, so this isn't super extensive. You hopefully bought this big thing because you thought that 200 resolution in ion mobility was probably an exaggeration, but it would be killer if it wasn't.
What else? Oh yeah! How to process the data? That's easy. Use basically anything you want.
Things that can do quan on MGF spectra?
Proteome Discoverer (including the free one).
SearchGUI + PeptideShaker + Reporter (this is still in beta).
Fragpipe will complain about it with popups, but I think you can just lie to it, say this isn't ion mobility, what are you talking about? it's just an MGF and you're fine. Can't say with 100% certainty, though.
A couple steps are necessary for Proteome Discoverer to process right.
You do need to use your spectrum selector so it knows what these files are. I suggest you lie and say the files are FTICR. This helps you keep all of your decimals places.
Tell it that unrecognized fragmentations are CID (honesty this fragmentation looks more like HCD and the two things are basically the same, so do what you want here) and give it your MS1/MS2 resolutions.
Big thing here if you are using MSAmanda, go under advanced in the node, scroll allllll the way to the bottom and max out your number of MS2 spectra per search (50k) you might have 200k MS2 per file. At 10k a piece that's a lot of searches. I recommend percolating. I also highly recommend using the most specific database that you can. It isn't a stretch to say that you might be giving Percolator e7 spectra to look at. If this is is RBCs do you really need all those mitochondrial peptides for all your tools to fuss over?
Recommended mass accuracy? -- If you've recalibrated your MGFs with MSFragger (crap. maybe I should write up a real tutorial. More later maybe, but MSFragger can recalibrate your spectra in the high mass range and it's AMAZING) -- 15ppm MS1 and 25ppm MS2/0.02 Da will work great. If you haven't calibrated I like 30ppm MS1 and 0.05 Da MS2. If you haven't calibrated in a month...?...go calibrate it.
Quan on one hit wonders might be wonkier than usual, but if you look close you'll see it probably isn't quan. It is that your FDR filters can let through a few more bad hits than you're used to. More on this later, but spectral libraries are AWESOME for TIMSTOF data. With the multithreaded MSPepSearch you can knock out a TIMSTOF MGF in like 90 seconds, no problem, but I think that the Orbitrap generated spectral libraries might not be as awesome as ones developed from TIMSTOF data.
Tuesday, February 22, 2022
Internal FAIMS Stepping for Top Down Proteomics!
I thought for sure I'd already posted this paper, but I can't find it!
Either way, this is worth talking about (again?). The basic idea is that if you really think about your populations of intact proteoforms in your samples in terms of:
1) What proteoforms you can actually detect (don't worry too much about Titin for now)
2) How much easier it is to identify smaller protein/proteoforms in most experiments (small proteins may need no microscans or scan averaging, while larger ones will need more)
3) How FAIMS compensation voltages can help enrich proteins of certain sizes (this was new to me and why I really like this paper)
4) How chromatography can also do some of #3
You can get some dramatic increases in identification rates versus both systems without FAIMS and systems where less FAIMS method optimization has been employed.
How much more?
Monday, February 21, 2022
Sample prep for tears from a common clinical test strip!
I guess the moral of this story is maybe we do sometimes need to invent new sample processing methods. This is, however, increasingly rare in my mind, but this seems to be a case because it appears that most people don't like having pipette tips near their eyeballs.
Friday, February 18, 2022
Tutorial --the easy way to set up pasefPRMs!
We just had to go through this in our weekly proteomics group meeting so it is fresh in my mind. I'll move fast before it fades. This way of doing pasefPRM is exclusively for the TIMSControl version above.
Even if you just got the ACS paper that came out 2 weeks ago on pasefPRM, if your TIMS control has been updated since ASMS2021 (Halloween 2021) you'll find things are slightly different.
You can build your target list exclusively through Skyline if you want. Go right ahead. That's awesome.
OR you can get your TIMSControl updated, process some DDA data through MSFragger or MaxQuant and then just build your targeted list like you would on another family of instruments you might be more familiar with. Here's how I do it with MSFragger.
I'm going to dig through a K562 digest for examples (click to expand) but I basically need my 1/k0, my observed peptide m/z charge and my retention time in seconds. I get these values from the PSM.tsv file from FragPipe generated results.
And this is why you want the newest TIMSControl:
You get a target editor! You can punch all these in, or you can make a CSV file and import it (way faster).
Obviously this is a very simple example I just made up, but you get these two visualizations that help you know if you're staying within the effective capabilities of the instrument. I purposely stacked two peptides at 32 minutes with different IMS ramps (highlighted in orange) just to show what that looks like. The image will scale as you load more targets and will provide a pretty intuitive visual way of seeing that is happening at each point in your experiment. There is obviously an upper limit for what you can target as you move across ion mobility ramps and optimizing that vs the MSinterval will be something you'll have to think about as you increase the number of targets, but otherwise you're good to go!
Thursday, February 17, 2022
Death to LCMS -- Nanopore proteomics is building inertia!
The last time I used a headline like that the one above there was a spike in blog traffic. I notice spikes in blog visitors because Google tries to talk me into activating ads whenever the traffic gets high. Pass. Thanks for the free hosting for 11 years, Mr. Google!
However, there is no doubt that alternative proteomics technologies are gaining momentum and the way the diminutive NanoPore (maybe I have to turn the ads on to get a spell check? I might have made up a word just now) is disrupting the genomics market, it would be silly not to pay attention.
This new study is even more evidence!
Phosphomatics! A whole suite of innovative tools for phosphoproteomics!
Big shoutout to one of the many Ben's of Proteomics (there is an informal club), Dr. Schulz, for the heads up on this great resource!
You can read about it here if you want, but you might want to just go to the link above and click around.
Wednesday, February 16, 2022
A practical guide to visualizing proteomics data!
Tuesday, February 15, 2022
Is it time for SUPER MASS SPECTROMETERS?
Does the thought of having the same ion beam connected to multiple mass specs induces "oh my god, I can't even get Waters to return my calls because I wanted to put it on the front of a beautiful new SCIEX (just a random example, but one where on Monday I'm just as randomly installing it myself)" level terror?
Or does the potential to split a single ion beam with the same efficiency (?? didn't make sense to me, but I've been asking people who know this stuff and it sounds like this company found a huge gap in efficiency in electrospray ionization and a truly unique way to work around it!!) make you start thinking of
what you could do with a super mass spec?!? If you hate HPLCs as much as I hate HPLCs, it's at least worth looking at. More details are available at www.tracematters.com
Monday, February 14, 2022
Top-down FPOP?!?!
Well....on the list of things that I'm glad no one has asked me to do today and I would probably have laughed and said "...ask me again in 10 years..." I present --
I mostly know about Fast Photochemical Oxidation of Proteins because the Queen of FPOP, Dr. Lisa Jones is right downtown doing crazy amazing protein structural stuff IN LIVING ORGANISMS with the technique. Or was, because Jones lab is leaving the world's greatest city and is heading to crummy UC San Diego. (Congratulations, UCSD!)
FPOP can induce a lot of different alterations on the exposed protein regions while largely not affecting any parts of the protein that are folded in such a way that they are on the inside of the protein and can't be attacked by these oxidation events.
This is a tough technique for bottom up and special software exists just for this technique. But top down? Which is already hard enough??
How do you get that data? They chopped up the DNA so it was off of their modified protein and they used a 15(!!) TESLA magnet for top down with CID and EAD. They also did bottom up traditional FPOP style so they had a good lead on the mods.
Sunday, February 13, 2022
12-Plex derivatization for accurate multiplexed quantification of citrullination!
Protein or peptide citrullination is a really important thing to some people and it's something that, in general, LCMS based proteomics is not very good at accurately identifying.
A citrulline is essentially an arginine plus 1.something very small (it's a Saturday and I don't remember it off the top of my head and I ain't looking it up). Even on an amazing Orbitrap running at 140,000 resolution once you get up to a peptide m/z of 800 or so it's tough for most search engines to determine what's a peptide that ends in a citrulline and what's the naturally occuing M+1 isotope.
You do get some hints in the fact that most trypsins won't cut a citrulline very well, but that's controversial as some of the super trypsins have been shown to do okay. You can also use the beautiful diagnostic fragment ion that the ProteomeTools project characterized. Since you can directly utilize that diagnostic ion in MaxQuant (or MSFragger) they're better tools to use.
What if you didn't have to guess and you could just multiplex citrullinated? Maybe that's worth derivatizing and...SCX...
Saturday, February 12, 2022
Ion parking + Infrared SPS MS3 of reporter ions = Big boosts in reporter signal!
Holy cow. I have to run, but there is so much to learn in this one.
One -- reporter fragmentation is an issue in HCD? Wait. What?
Two -- I thought Syka retired, good to see him on this paper! Or it was the other guy that I always get mixed up with him? I forget.
Three -- HCD performance is m/z related? Did you know this? Should I pretend that I did before this? Meh.
What's all this add up to if you hack up an instrument to park ions and then fragment them in a way that isn't m/z dependent?
2.39x increase in average reporter signal!! For anyone at those limits where 2x would be transformative. This could be a really big deal in 3 years when it's commercially available (just made up a number).Friday, February 11, 2022
APCI and APPI -- for identifying peptides???
Thursday, February 10, 2022
Saturday, February 5, 2022
Ancient viral insertions in our DNA present an entirely new set of potential targets for immunotherapy!
Friday, February 4, 2022
SMACK! An example of how to listen to your mass spectrometers!
Are your mass specs trying to tell you something? For my stuff, my current favorite is RawBeans but that's for -omics and I can't imagine it's current iterations would translate well to your high throughput validated targeted assays.
SMACK is an open toolkit that thinks through all the hard stuff that is pretty convenient to not know about when you're out there throwing around words like "translational" about your work. This group is on the ground applying new mass spec assays to patient samples and has to think about these things, and it's beyond cool that they'd make these tools open-source. You can drop $250k on setup plus $40k/year on LIMS systems that don't have trend monitoring capabilities nearly this advanced.
Thursday, February 3, 2022
Contribute your thoughts to a labcoat designed for labs with this 90 second survey!
This is quite possibly the best description of a labcoat ever. This group is trying to get 1,000 scientists to complete a brief survey so they build a labcoat that makes sense for people in a lab. It popped up this morning on r/biochemistry. You can find the article and link to the survey here.
Wednesday, February 2, 2022
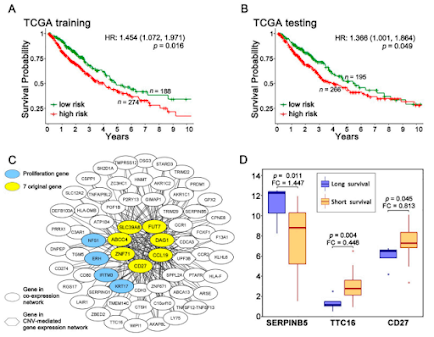
Inspirational multi-omics of lung cancer recurrence without ever going to a lab bench!
Some of the biggest and most expensive projects in our field are just building big resources. I really like this study despite the statistics and cancer terminology heavy writing (with a lot of probably necessary all capital letters?) because is shows how people can put these to use and find cool biomarkers(?) without leaving their house.
This group in a beautiful couch burning mountain city (I was there for that one. It was nuts. Turns out that's just what they do there. Find a nickel on the sidewalk? Get the gasoline and the recliner! Beat a historic rival for the first time in 412 years? People will still talk about it almost 20 years later! It wasn't the best day to be wearing my school colors which, as a super bright orange can still be seen by drunk kids even when there is tons of smoke) pulled out the stops and dug into at least 6, maybe 8, cancer resources, including the most recent release of the Cancer Cell Line Encyclopedia Proteomics Project which might have gotten a facelift? It looks super sharp.What really stands out here is that this group didn't just fish around looking for what stood out as intense and obviously changing in abundance. They went into these massive resources with what seems like a pretty clear hypothesis that centered on 7 genes previous results from their lab indicate are linked to cancer recurrence. By thinking about how each of these resources, which are human samples, so intrinsically flawed since we can't control all the variables in an actual tumor or tumor cohort resource, in a medically relevant sense they can continually refine their hypotheses. Even if you can't follow their full line of thinking here (which I can't, I've already went to Wikipedia 3 times) it seems like a great story that leverages what information you can get from CCLE and the Achilles KO project to support or shoot down observations from much older microarray and RNASeq analyses of cancer projects.
They do end up going to the bench and doing some western blots to back up what survived from their initial hypothesis and it looks like they're onto something.
So, if you're out there trudging through your one millionth pulldown experiment because your boss got the money to capture every protein-protein interaction of some sea slug, or (maybe more pertinent) you are doing 46 offline fractions of each tumor and there are two -80C freezers of tumors left to go maybe you should check out this paper. I bet it is easy to stop and wonder if maybe you're doing a lot of work to make a resource that no one will ever use, right? But I think this shows that, if you trudge through and keep doing it right, you might be enabling some smart people out there in the world to do important things with it, and really, is there a better use of that great big noisy vacuum chamber?
Tuesday, February 1, 2022
BioSaur -- Get all the peptides with ion mobility support!
There are a lot of warnings on the preprint from the publisher so I can't grab pictures. Biosaur looks like an extremely promising new toolkit for people out there wrangling with FAIMS and TIMS and maybe other ion mobility front end things!
You can get it at this Github (the preprint hyperlink includes the period at the end of the sentence so it will look like the link is down, just delete the period).
And you can directly open the PDF for the preprint from this link.