Tuesday, July 28, 2015
Process DIA data directly in Proteome Discoverer 2.0 with DIA Umpire
Alright, this DIA stuff is confusing. There are methods all over the place. WiSIMDIA on the Fusion, pSMART, multiplex-DIA, and even boring old SWATH. Software-wise, there is tons of stuff out there. This weekend I processed some DIA data from a Fusion directly through Proteome Discoverer 2.0...and it looks amazing.
The data in question was ran through the DIA-Umpire to convert the data into a handy-dandy MGF file format. You can find details on the DIA-Umpire in this Nature Methods paper by Chih-Chiang Tsou et al., out of Alexey Nezvizhskii's lab. Essentially, it is a (currently) command line driven program that takes your DIA data and "deconvolutes" it down to a format that is friendly to the proteomics processing pipelines we already know and trust. How does it work? No idea. But it works, and the data looks amazing (did I say that once already?)
Here is a random high scoring PSM I grabbed. Looks pretty incredible, right? They all do. And I didn't have to change my workflow at all. I used SequestHT, target decoy and my normal basic consensus report. I ended up with a ton of IDs and really nice true FDRs at every level I set them at (PSM, peptide, and protein).
If you are interested in identifying peptides via DIA and you are a little swamped by your software options, you might want to check this out. I'm tired of learning new software interfaces -- lets put everything in Discoverer!
Monday, July 27, 2015
Another amazing Boston trip!
Yesterday, I somehow suckered around 80 people into spending about a whole day in a big room talking with me about Proteome Discoverer 2.0. We ran through a lot of different processing ideas and scenarios and I got a ton of feedback to pass on to the Proteome Discoverer team (which I should probably be doing right now rather than blogging this...oh well...it WILL be passed on shortly)
Today was more fun of popping in to see a bunch of different labs and talk about different processing needs and how we can address them with Proteome Discoverer. Now I've got several cool puzzles to work on (man, everybody is doing something cool in this town!!!). Anyway, no real news here, just a shout out to the great people of Boston/Cambridge for their time and energy in making this (from my standpoint) a fantastically (that's a word?) productive trip!
Sunday, July 26, 2015
Want to take a full intensive course on targeted proteomics online?
This is why I love Twitter for learning stuff! So much good information out there -- such as this 21-part course on targeted proteomics put on recently by a ton of experts in targeted proteomics in Zurich.
You can watch all the videos here!
Shoutout to Ben Collins for leading me to this list of all the videos in order!
Friday, July 24, 2015
Outstanding review on the evolution of the Orbitrap
The Orbitrap hasn't been around all that long, yet there are tons of different flavors. A really fun aspect of my job is that I sometimes get to go into a lab where someone has been running an Orbitrap XL (which is an awesome instrumet, btw!!) and I get to be there when they get to see what their new QE HF is capable of.
Can you still get the same data out of your Orbitrap XL? In a lot of cases, hell yes you can! Can you get that same information on the QE HF in...one-quarter or one-tenth the time....sometimes the answer is a resounding yes.
So what are the differences? For an incredibly thorough (and very pretty) review of where the Orbitrap was, is, and maybe will be next check this paper from Shannon Eliuk and some guy named Makarov?
Its open access and a great read!
Thursday, July 23, 2015
Does DMSO addition affect label free quantification?
DMSO as an additive for nanoLC proteomics applications is still pretty polarizing. If you want my opinion on it, it is: yes, you get more signal and peptide IDs, but you should anticipate requiring more cleaning and maintenance on your instrument. If you don't mind the downtime and the signal intensity is paramount...well, that's your choice. For me? I would run DMSO if I was doing experiments on your instrument, but I wouldn't use it on mine...
To further investigate the affects of DMSO as an additive Dominika Strzelecka et al., checked to see if adding 3% DMSO would affect their label free quantification. I stole the figure above from the open access paper. In A, you see the ID'ed peptides. In B you see the quantified peptides. In the end they found that DMSO really didn't affect the quality of the quantification, though maybe the increase in signal does help you quantify more.
Me? I'm most interested in the shift in identified peptides!?! Out of ~2500 peptides ID'ed over 1000 were differentially ID'ed by changing the buffers!
Highly recommended paper that adds more info to a very interesting topic.
Wednesday, July 22, 2015
Proteomics in negative mode!
Wow! What is happening in this picture? Something flat out crazy and awesome.
The paper is in press at MCP here from Nicholas Riley et al., out of some guy named Josh Coon's lab.
What is it? A hacked LTQ Orbitrap with a new collision source. They call it the "Multipurpose Dissociation Cell" and it massively improves the signal, speed, and utility of ETD fragmentation. It improves it to the point that it makes doing proteomics completely in negative mode something that everyone has thought about something that is actually a possibility.
Just last week I spent some time explaining why we don't do negative proteomics for PTMs: poor ionization, fragmentation sucks, there aren't good tools for translating the data, etc., Amazing how this field evolves!
To get around poor ionization, this study switched the buffers around. High pH reverse phase!
To get good fragmentation, they use their awesome new MDC source to perform Acitive Ion Negative ETD (AI-NETD)
To do the data processing, they studied the AI-NETD fragmentation spectra and determined the new charge-reduction loss masses and processed the data to remove these components. Then they used a modified version of OMSSA that was set to read a(dot) and x fragment ions.
I had to pull the Wikipedia peptide fragmentation chart to figure out where things are coming apart. Wow, right?
This isn't the first run at doing negative proteomics. It isn't even the first run at doing negative ETD fragmentation. This is, however, the first time that we've seen this approach produce results on the same scale as we get from positive fragmentation studies.
In one run they broke the 1,000 proteins ID'ed barrier on yeast.
Using multiple enzymes they come close to 100% coverage of the yeast proteome. How's that for on the right scale!?!
But that isn't really where the power of this approach is needed. We can get whole proteome shotgun coverage. We've kind of got that one in the bag. This opens up a whole new capacity for ions that prefer negative charges. Like many post translational modifications do. Heck, this might even open us up for more high throughput analyses of completely different biomolecules like oligonucleotides.
I want to extend a personal thank you to the authors here. I was trying to come up with reasons to get out of bed early this morning and just when "staying employed" didn't seem like it was enough to tip the scales, I found this in MCP. Now I'm up, motivated and about to head out the door to see what else all you brilliant people are up to out there!
Tuesday, July 21, 2015
Added PRM/DIA to QE cycle time calculator
Okay! So that got downloaded a LOT!!! Thanks for the feedback!
Motivated me to do some updates on my lunch break. In the Q Exactive Family Cycle Time calculator you will find some important updates including an important "read me" section including my assumptions and references for where I got the math I used.
More importantly, however, is that I added sections of parallel reaction monitoring calculations (PRM, also called T-MS2 in QE Tune previous to version 2.4) and simple DIA.
You can get the new and improved calculator here.
Monday, July 20, 2015
Q Exactive/Plus/HF cycle time calculator!
I got kind of motivated Sunday and finally knocked out a project that I've wanted to for a while.
Introducing the Q Exactive TopN cycle time calculator. This simple Excel spreadsheet allows you to pull down the resolution and insert the max fill time for your method and will give you the minimum and maximium possible cycle times for your experiment.
Its for estimation purposes only but its something I like not having to think about. You can download it here.
BTW, in this exercise I'm defining "cycle time" as something like: "the amount of time it takes to perform all of the events you asked the instrument to do before it goes back and starts at the beginning (in this case, start the next MS1 scan)"
I hope to expand it later to include targeted and DIA experiments but it might be a bit. Oh, and as always this isn't any official vendor type stuff (see dislcaimer section in spreadsheet AND this blog and please don't sue me!)
Sunday, July 19, 2015
How do I determine my peak width for optimal dynamic exclusion settings?
Whenever we talk about ideal dynamic exclusion settings I always start saying things like setting your numbers according to your peak widths. This week I realized that not everyone knows how to do this!
The way I do this is by opening a file in Xcalibur. Once I've done that I set some ranges with tight extraction windows so that I can look at a couple of good peptides. If its a human sample I look for two specific peptides. Their m/z(s) are:
722.32477
and
756.42590
These are two peptides from albumin. The first is relatively hydrophilic and the second is relatively hydrophobic. Since albumin is everywhere I'm going to always see them, whether its plasma or cell pellets or whatever.
To extract the peaks, click the thumbtack on your chromatogram. Next, right click anywhere on the chromatogram. Before you do anything else, toggle the tab over to the "Automatic processing" tab.
Set up the mass tolerance settings as above. I like 5ppm, though you can probably go lower on a recently calibrated Orbitrap, but 5ppm will do it. I don't use any smoothing or nuthin unless I'm trying to prepare a figure for people outside of the field to look at.
Next toggle back to ranges. You'll need to set it up as so:
So, add 2 ranges to your chromatogram tabs. Set them as base peaks and so that they only look at the MS1 scan filter. The box above shows how to search for the hydrophilic peptide. Repeat for the next row but enter the range(s) for the mass of the hydrophobic peptide. Hit "OK". You should now be looking at something like this:
You can't see much until you actually zoom in on the peak. You do this by clicking and dragging across the peak individually. On this 2 hour gradient the first one looks like this:
Now, we can just eyeball this and say "Yup! Thats about 30 seconds" or we can drag the mouse across the line. Normally in chromatography we would consider the peak width by measured across the peak at half height. This is NOT how we do this for dynamic exclusion. What you want to do is measure the peak from the intensity threshold cutoff in your method (or as close as you can get!)
(click to expand if you want to see it!
When this ion was first detected within this mass range on the instrument its intensity was 2e5. On a Q Exactive, my intensity threshold cutoff filter is typically in the 1-2e4 range. So the first time that this ion appeared in the chromatography it could be selected for fragmention. If it was then the dynamic exclusion would start counting at that point. In this case it was at 57.71 minutes. If I look at the far end of the peak it disappears around that range and it is just about exactly 0.5 minutes after it started. So this is a 30 second wide peak. Repeat for the second peptide.
It is important to keep in mind that some peaks will look different than others. In the case of these two peptides, I expect the second one to be wider than the first. From threshold to threshold on a 2 hour gradient it is 45s to 55s. Why is this? No idea. Not a chromatographer, but I can only set one value for my dynamic exclusion. If I'm a soloist I'm going to err for the higher value. If I'm a two-timer (description of my terms for this here) I've got to be a little more intelligent and I might want to look at 4 or 5 more peaks to get a good idea of what my real peptide elutions peaks look like so I get the ideal number of MS/MS events.
Saturday, July 18, 2015
Need another search algorithm? Check out ProLucid!
You know what the world needs? Another awesome search algorithm! Prolucid is a brand new engine from the Yates lab that is described in this open access new paper. (oh...and I don't think that is the logo for it, I think that is something else...)
Why would we download it and give it a shot?
More peptides, of course!
If you don't want to check out all the math in the paper you can just download it here.
As a side note, have you seen this Wikipedia page that lists mass spec software? It definitely misses some things but its pretty nice.
Friday, July 17, 2015
De novo sequencing workshop at CSHL
I was lucky enough to get an invite again this year to help out with the Cold Spring Harbor Labs proteomics course. Great group and great instructors (and great data coming off the Q Exactive Plus!) If you are on the outside and want to get into this field I don't know of any better program in the country. Today's highlight is Karl Clauser's half day de novo workshop.
Sad you can't take this fun course? Don't be cause you can download the whole thing here!
Thursday, July 16, 2015
Native protein extraction for tryptic digestion
When we harvest out our proteins for shotgun proteomics one of the first things we do is mess them all up. The proteins that get to meet our trypsin are completely inactivated by reduction and alkylation and whatever other awful things we do to them. Is there a chance this is causing problems in re-assembling the original biological picture?
No, wait, maybe we will!
Avantika Dhabaria et al., may even have a mechanism for us to figure it out! In this new paper in JPR, they introduce us to: "A high-efficiency cellular extraction system for biological proteomics!" Wait...I added the exclamation point. Holy cow. There are exclamation points everywhere! Google's analytics engine just informed me that I use those more than periods...TIME FOR MORE ELLIPSES!...
Anyway, they used acoustic pulses and all sorts of other things and compare the samples. Is this better for us downstream? Who knows! But if you have a mechanism where you should be investigating functional proteins....then you have a protocol!!
Wednesday, July 15, 2015
Special enrichment strategies!
What if I came to your lab and said that I needed to see every bit of every protein in a sample? I bet you'd 1) think I was crazy and 2) would have some pretty good ideas for finding peptides that you normally miss.
We compromise because we know we have to. Not everything sticks well to C-18. Good peptides are below and above the mass range of our instrument settings. Molecular weight cutoff filters like we use in FASP lose some cool small signaling molecules. Just a few examples. I'm sure you can come up with others.
But if you really had to do it, how low could you go? Great question, right? And that's why we have awesome projects like the C-HPP. A brand new chapter from this endeavor is in this month's JPR (here!)
In this study they do this...
Yeah....glad it was them and not me. That is a LOOOOONNNNGGGG day (semester?) of wet bench work, not to mention instrument run time.
How'd they do? ~9000 proteins. Not too shabby. But that's not even the goal. The goal was trying to hunt down missing proteins. Things that look like they should be expressed from what we know of the chromosome and exons/introns and what not. And they found at least 30 of them! One step closer to the complete proteome!
Tuesday, July 14, 2015
Sleep deprivation changes visible at the genome level!
I'm not calling anybody out or anything but there are a lot of people in this field that don't seem to sleep very much. Maybe its a bias toward the people that I know, but I feel like if I'm stuck on something at 2 or 3am I've probably got a pretty good chance of getting in touch with somebody. Heck, maybe I've got a better chance than I do at 3pm.
This interesting study at Uppsala (described in this simple blurb here) took a look at what happens if we skip a full night's sleep and the results are pretty interesting. After skipping just one night of sleep you can see differential regulation at the genetic level. Not something rapid like the phospho- or kinome level, but actually genetic abundance issues.
Now there's always this question for genetic abundance changes -- does this really translate to anything at the protein abundance level? Might be an interesting follow-up for someone (hint?).
Sunday, July 12, 2015
PD 2.0 workshop for Cambridge!
Hey Cambridgians and Bosticians! Lets waste a day away talking about the awesome power of Proteome Discoverer 2.0.
This is an interactive workshop. Intro slides for maybe an hour and then live demonstration. We'll start with easy simple workflows and work our way up to crazy stuff. Along the way I'll take notes on your comments/suggestions and pass your feedback onto the programmers and PD team.
If we have time I'll show you what new special powers are coming in the next version.
If you want come, email Susan.Bird@ThermoFisher.com
Saturday, July 11, 2015
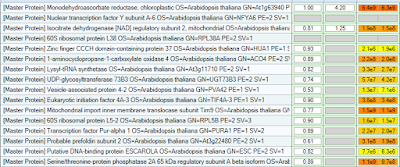
Outstanding review on proteogenomics!
Proteogenomics is some great stuff. But it isn't easy stuff yet. You might notice from recent blog posts that its starting to become a bit of an obsession of mine... The promise is there. The data is there, but integrating RNA-Seq data with Proteomics is still no easy task.
For an overview of where we are (as of 2014) you should check out this great review in Nature Methods from Alexey Nesvizhskii. It is open access, remarkably clear, and very thorough.
Friday, July 10, 2015
Thursday, July 9, 2015
Reporter ion proteomics probes morphine tolerance and addiction
I love the city of Baltimore. It has its problems, though. Chief among them might be our new title as "the heroin capital of the US..." There is obviously some sort of a link between this fact and our extremely high crime rates. Opiates like morphine and heroin, though, are just about ubiquitously abused throughout the world and sinister everywhere.
To gain insight into the function of these abused substances, Steve Stockton et al., used an elegant quantitative approach to study the effects of morphine on subcellular fractions from synaptic cells. After a lengthy and thorough sample preparation technique they used iTRAQ on an Orbitrap Velos to find proteins disregulated by the drug.
Now, I'm no neuroscientist, and I'm not going to pretend I understand what they found and its impact on their field, but anything that gets us closer to understanding opiate tolerance and addiction gets categorized under AWESOME in my book. I really can only assess the methodology and they did a nice job. The paper is currently in press at MCP and open access and you can find it here.
Wednesday, July 8, 2015
How to sort your quantification report by individual ratios in PD 2.0
This question has come up a couple of times. So I have these nice ratios now. But what if I want to sort by one of them. I can click and click and I can't sort by individual ratios.
You need to change one thing in the Advanced Tab in your Consensus report:
Create Separate quan columns in your Peptide and Protein Quantifier. Now you'll get a completely separate column for the Ratio counts and variance for that channel. And you can sort by any one that you want to.
Tuesday, July 7, 2015
Google reports BRIMS portal hacking -- appears to be in error.
So...Google appears to be getting a little more aggressive about sites that it can't "crawl" over freely. The newest one is this suggestion that the BRIMs portal may be hacked. Keep in mind that this is an automated method that looks for changes in what it can see of a website. We have received no indication that there is a problem with the site (the Thermo Omics software manager is one of the people sitting at a table with me right now and I think they'd tell him...)
Monday, July 6, 2015
Intro to proteomics workshop in October!
From the inside its easy to forget how daunting proteomics is.
One talk I saw a slide from recently referred to "the tyranny of the mass spectrometer". If you're on the outside its hard to know what questions to ask and what is easy to do with the technology vs what is hard. For those of us on the inside, having a more knowledgeable customer base or collaborators only serves to make our lives easier.
Whats the solution? More education! And workshops geared toward researchers who want to know:
1) What can the MODERN mass spectrometer do
2) How can it help me to do it
3) What is a reasonable experiment vs. what is an extremely difficult one.
You can register for this one in Germany here. What we need is a bunch of these in every country!
Sunday, July 5, 2015
N15 labeling quantification in Proteome Discoverer 2.0
I'll not lie to you. This was on my "to-do" list since December and I didn't think I'd get to it until next December but I lucked into a couple spare hours on Friday and here we go.
Without further ado: how to do N15 quantification in Proteome Discoverer 2.0. This is similar to how my old tutorial went for PD 1.3. (BTW, there is an alternative way of doing this in PD 2.0 that I just figured out, but I think this one will be the cleanest! Lets see!)
Go to Administration > Maintain Chemical Modifications. And add these modifications (click to expand):
If you want to add it to an existing Study you will now need to save that study, close it and re-open it for this method to work. Better yet? Just close any open studies. I'm not done yet.
Go to Quantification methods (still in Administration). Add a new Quantification method. You can write it from scratch OR you can clone it from a different method. Here I'm cloning it from the Full 18O method:
Alright...this is where it gets complicated. Different amino acids can integrate different amounts of N15. Some only have one N, while others have up to 4. You need to create 4 labels, one for each possibility. It ought to look something like this:
Next...well...you need a good dataset to test it on. If you are interested in this topic its likely that you already have one. I...didn't. Fortunately, however, I found a fantastic study from a couple of years ago from H. Zhang et. al., out of Utreckt. If you want direct access to the data it is PRIDE dataset PXD000177 (direct link? not sure if you can direct link into PRIDE due to user account stuff).
Set up your workflow the way you usually would (I used the pre-made SILAC template) but add all of your modifications:
Set up your Consensus as normal. Just make sure the "Peptide and Protein Quantifier" node is involved. If it isn't you should get a little pop-up to remind you, but its better to just do it yourself :)
Note: there is a slightly easier way around this on newer instruments. Since this is an MS1-centric quantification method you don't actually have to have fragmentation of both your light and heavy species to get quantification. In fact, you can set your instrument to only fragment your light species (or heavy) then you don't have to use data intensive dynamic mods for your searching. Just a thought. Here I'm just doing both.
I'm going to have a coffee while Percolator digs through 27 dynamic modifications...actually, it wasn't that bad...and I appear to have packed all the coffee (moving is AWESOME!).
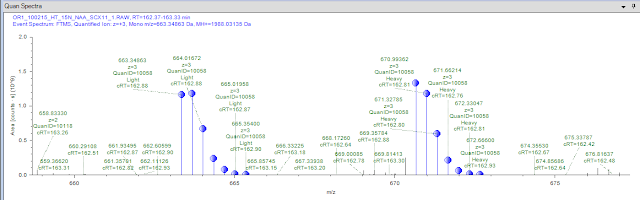
Okay, here is the $64 question: Did it work? Yeah, I think it did...
I ran one SCX fraction from this study. They did extensive SCX fractionation. Only in 2 cases did I have 2 peptides from the same protein identified (hence the missing ratio standar errors). But I generated ratios...and...
...I get quantification spectra that seem to make sense. This peptide was found as a triply charged heavy and light and looks pretty believable. And crude math in my head according to the length of the proteins makes this 24 Da shift between light and heavy seem perfect to the peptide sequence.
I'll try to download one of the full datasets from this study and search the data the same way they did and see if I can match their results. Now...this needs said... This method isn't perfect. There are better ways of doing this experiment (check out the awesome software package written by Princeton and mentioned in an earlier blog post) but PD can do it. And if you really want to do it this way you really can iron out the rest of the bugs. But this ought to help you get started. I'll try to put together a video for this when I have time.
Without further ado: how to do N15 quantification in Proteome Discoverer 2.0. This is similar to how my old tutorial went for PD 1.3. (BTW, there is an alternative way of doing this in PD 2.0 that I just figured out, but I think this one will be the cleanest! Lets see!)
Go to Administration > Maintain Chemical Modifications. And add these modifications (click to expand):
If you want to add it to an existing Study you will now need to save that study, close it and re-open it for this method to work. Better yet? Just close any open studies. I'm not done yet.
Go to Quantification methods (still in Administration). Add a new Quantification method. You can write it from scratch OR you can clone it from a different method. Here I'm cloning it from the Full 18O method:
Alright...this is where it gets complicated. Different amino acids can integrate different amounts of N15. Some only have one N, while others have up to 4. You need to create 4 labels, one for each possibility. It ought to look something like this:
Next...well...you need a good dataset to test it on. If you are interested in this topic its likely that you already have one. I...didn't. Fortunately, however, I found a fantastic study from a couple of years ago from H. Zhang et. al., out of Utreckt. If you want direct access to the data it is PRIDE dataset PXD000177 (direct link? not sure if you can direct link into PRIDE due to user account stuff).
Set up your workflow the way you usually would (I used the pre-made SILAC template) but add all of your modifications:
Set up your Consensus as normal. Just make sure the "Peptide and Protein Quantifier" node is involved. If it isn't you should get a little pop-up to remind you, but its better to just do it yourself :)
Note: there is a slightly easier way around this on newer instruments. Since this is an MS1-centric quantification method you don't actually have to have fragmentation of both your light and heavy species to get quantification. In fact, you can set your instrument to only fragment your light species (or heavy) then you don't have to use data intensive dynamic mods for your searching. Just a thought. Here I'm just doing both.
I'm going to have a coffee while Percolator digs through 27 dynamic modifications...actually, it wasn't that bad...and I appear to have packed all the coffee (moving is AWESOME!).
Okay, here is the $64 question: Did it work? Yeah, I think it did...
I ran one SCX fraction from this study. They did extensive SCX fractionation. Only in 2 cases did I have 2 peptides from the same protein identified (hence the missing ratio standar errors). But I generated ratios...and...
...I get quantification spectra that seem to make sense. This peptide was found as a triply charged heavy and light and looks pretty believable. And crude math in my head according to the length of the proteins makes this 24 Da shift between light and heavy seem perfect to the peptide sequence.
I'll try to download one of the full datasets from this study and search the data the same way they did and see if I can match their results. Now...this needs said... This method isn't perfect. There are better ways of doing this experiment (check out the awesome software package written by Princeton and mentioned in an earlier blog post) but PD can do it. And if you really want to do it this way you really can iron out the rest of the bugs. But this ought to help you get started. I'll try to put together a video for this when I have time.
Saturday, July 4, 2015
customProDB -- RNA Seq to FASTA
Proteogenomics, how I yearn to find your true potential in a push-button sort of way!
This is NOT a bush-button sort of way, but its a solid tool. customProDB is a couple of years old and is the work of Xiaojing Wang and Bing Zhang out of Vanderbilt. You can read the open access text here.
What you need to use customProDB? A pretty good understanding of a recent version of R, BioConductor and a couple other add-ins:
These are the details taken from the customProDB BioConductor page here.
Not push-button, but very powerful.
Do you have an RNA-Seq to FASTA conversion tool that is push-button? Want a ringing endorsement and maybe a bunch of rapid citations for your tool? Email me or add a comment and I'll enthusiastically check it out!
Friday, July 3, 2015
ETD Supplemental Activation vs EThcD!
ETD is awesome. Is it complicated? Yeah, a little, but on the right sample and the right conditions it gives you data you can't get from anything else.
In CID or HCD fragmentation you only have two things to think about: How many ions I want to fragment and what collision energy (CE or nCE) that you want to use.
ETD is a little more complicated. On top of how many ions you want to fragment you need to think about:
1) How much ETD anion to use
2) How long to allow the ions you want to fragment to react with the ETD anion
Number 1 looks more complicated than that because you have another time that you need to use. This is called the ETD injection time. This is the same as the AGC target for your ion you want to target. In other words, you tell the instrument "use 5e5 ETD anions OR spend this amount of time trying." This is really important on the last generation of instruments, such as the LTQ Orbitrap XL or Elite with ETD. The intensity of the anion produced by the filament-driven ETD source tends to drop over time (between cleaning and tuning cycles). You can compensate for that drop by allowing greater fill times to reach your target. It is virtually a non-issue with the Fusion and Lumos systems as the ETD production stays almost perfectly stable day after day after day.
But this isn't the topic of this blog post. This is:
This toggle was present (though it looked a little different!) on my first Orbitrap, the great XL plus ETD. There were no options for it. You either had a checkmark in the box or you didn't.
I think ETD Supplemental Activation (SA) was described in Swaney et. al, here (open access!). However, I think there was a paper by David Horn that pre-dates it that did something similar with ECD. A quick Google Scholar search doesn't pull it up for me, but I'm pretty sure that it exists.
Nevermind. In the Swaney paper you'll see the logic behind the use of Supplemental activation and its power. In a nutshell, you allow the ETD anion to react with your target peptide for the set reaction time.. and then you add just a little bit of CID (called CAD in the paper I'm referencing) to improve things a bit. This leads to an improved formation of the c and z ions produced by ETD. Awesome, right!?! There can, however, be some minor drawbacks. You can sometimes impart a 1 Da shift on some of the fragments. You can also sometimes induce secondary fragmentation events. On the earlier LTQ Orbitraps you couldn't control the amount of SA that you employ. It was set at some nice low level to keep secondary fragmentations rare. I think starting with the Orbitrap Elite (I get all these instruments mixed up) you have the ability to control how much SA you use. Too much and you start to see secondary fragmentation events. If your search engine isn't set up to handle these possible b/y or hybrid fragments you might lose data. Again, however, you should primarily be seeing c and z ions (with some mass shifts here and there).
This guy, however, is a whole 'nother animal. In EThcD you WANT to introduce secondary fragmentation events. Its the whole point. After ETD fragmentation is complete then you induce HCD "all-ion" fragmentation on the ETD fragment ions.
Correct me if I'm wrong, but I think this was first described by Christian Frese et. al., in this paper using a hacked Orbitrap Velos (I packed my filing cabinet and I don't have online access to this one).
A great reference for this technique is this poster by Protein Metrics. In this they show the power of this technique for PTMs. You get one scan with fragmentation data characterizing your PTM spread and your peptide spread. There are some killer studies out there using this technique for PTMs and proteins and bunches of other things. If you want to try this technique on just peptides, it is critical that your search engine is equipped to handle c/z/b/y ions to get good data. On Sequest you just add a weight to your ions. On Mascot (depending on the version) you may need to actually go to the server itself and change the configuration to allow Mascot to read all 4 fragment types into its calculations.
Thursday, July 2, 2015
Membrane proteomics -- a nice new review!
So much of the cool stuff in proteomics, particularly when we're dealing with drug and stress response relies on events occurring at the cell surface with signals transmitted through to the cytoplasm. Membrane proteomics is no easy task. The proteins have big hydrophobic regions that are hard to separate from the membrane chunk and sometimes even more difficult to digest and ionize.
If you are interested in this topic:
There have been tons of reviews on this topic (including my humble attempt to sum up membrane phosphoproteomics..insert shameless plug!). This new one in press at MCP from Xi Zhang is a whole lot better and much more up-to-date and is a stellar review for someone interested in this topic and the cutting edge technologies.
Notably, Dr. Zhang's review never mentions one of the classic (and still maybe one of the best!) solvent digestion methods pioneered by Josip Blonder and Dick Smith. If you're going into getting a ton of membrane coverage, I think it would be a mistake to forget about this one!
Wednesday, July 1, 2015
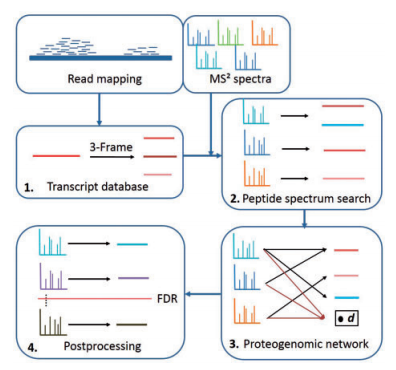
We know too much about biology to rely on 6 frame translation!
Everybody knows someone who has or is getting RNA-Seq data. And we've seen the awesome papers where using this data has given us new mutations and cleavage sites and things that we didn't get from a normal FASTA. Connecting those two, however, still seems to require a dedicated bioinformatician.
From my perspective I figure the easiest way to get these things set up is to change the RNA-Seq data into a FASTA that I can input into my normal proteomics pipelines (cause I already know how to use those).
Even this, however, isn't without problems. If you take the nucleotide sequences as they are you still have to change those into proteins. The simplest way to do that is to do a 6 frame translation.
Check out this example. We line up the nucleotides and pick 6 different starting points (here counting both DNA strands). We know that 3 nucleotides code together for one amino acid, but we don't know what our frame of reference is. Where do we start counting? In a 6 frame translation you go ahead and do them all.
In this nice example above, four of the possibilities can be pretty much ruled out because there are so many stop codons that they likely don't code for anything that we can sequence in a shotgun experiment. But ruling these out can get real tiresome real fast and a LOT of sequences aren't this clear cut. So you basically make a database that is 6x bigger than it needs to be with 5 sequences that aren't real (lets not even consider what this does to FDR. Lets not. Its late. And we aren't even talking about introns and exons, which is worse).
Franziska Zickmann and Bernhard Y. Renard think we should be smarter than this. And they have a solution that is better that they detail in this open access paper that they call MSProGene.
This whole packaged solution isn't exactly what I'm still wishing for (easy, intelligent FASTA generation with redundancy removal), but it shows how we can use the sequencing data to help us with another big problem -- protein inference! We can combine what we know from the genes present to help us figure out what proteins our peptides came from! MSProGene is a complete package that does the work from RNA-Seq and LC-MS data all the way to peptide and protein lists (its uses the MS-GF+ search engine).
While its still probably a tool aimed for bioinformaticians to work through combining these rich and complicated data types, it undeniably adds some great new tools and insight into this pipeline.
Subscribe to:
Posts (Atom)