I'll not lie to you. This was on my "to-do" list since December and I didn't think I'd get to it until next December but I lucked into a couple spare hours on Friday and here we go.
Without further ado: how to do N15 quantification in Proteome Discoverer 2.0. This is similar to how my old tutorial went for PD 1.3. (BTW, there is an alternative way of doing this in PD 2.0 that I just figured out, but I think this one will be the cleanest! Lets see!)
Go to Administration > Maintain Chemical Modifications. And add these modifications (click to expand):
If you want to add it to an existing Study you will now need to save that study, close it and re-open it for this method to work. Better yet? Just close any open studies. I'm not done yet.
Go to Quantification methods (still in Administration). Add a new Quantification method. You can write it from scratch OR you can clone it from a different method. Here I'm cloning it from the Full 18O method:
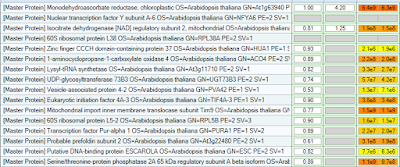
Alright...this is where it gets complicated. Different amino acids can integrate different amounts of N15. Some only have one N, while others have up to 4. You need to create 4 labels, one for each possibility. It ought to look something like this:
Next...well...you need a good dataset to test it on. If you are interested in this topic its likely that you already have one. I...didn't. Fortunately, however, I found a fantastic study from a couple of years ago from H. Zhang et. al., out of Utreckt. If you want direct access to the data it is PRIDE dataset PXD000177 (direct link? not sure if you can direct link into PRIDE due to user account stuff).
Set up your workflow the way you usually would (I used the pre-made SILAC template) but add all of your modifications:
Set up your Consensus as normal. Just make sure the "Peptide and Protein Quantifier" node is involved. If it isn't you should get a little pop-up to remind you, but its better to just do it yourself :)
Note: there is a slightly easier way around this on newer instruments. Since this is an MS1-centric quantification method you don't actually have to have fragmentation of both your light and heavy species to get quantification. In fact, you can set your instrument to only fragment your light species (or heavy) then you don't have to use data intensive dynamic mods for your searching. Just a thought. Here I'm just doing both.
I'm going to have a coffee while Percolator digs through 27 dynamic modifications...actually, it wasn't that bad...and I appear to have packed all the coffee (moving is AWESOME!).
Okay, here is the $64 question: Did it work? Yeah, I think it did...
I ran one SCX fraction from this study. They did extensive SCX fractionation. Only in 2 cases did I have 2 peptides from the same protein identified (hence the missing ratio standar errors). But I generated ratios...and...
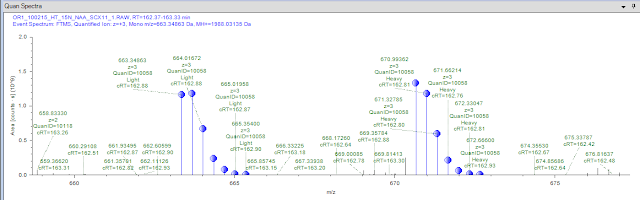
...I get quantification spectra that seem to make sense. This peptide was found as a triply charged heavy and light and looks pretty believable. And crude math in my head according to the length of the proteins makes this 24 Da shift between light and heavy seem perfect to the peptide sequence.
I'll try to download one of the full datasets from this study and search the data the same way they did and see if I can match their results. Now...this needs said... This method isn't perfect. There are better ways of doing this experiment (check out the awesome software package written by Princeton and mentioned in an earlier blog post) but PD can do it. And if you really want to do it this way you really can iron out the rest of the bugs. But this ought to help you get started. I'll try to put together a video for this when I have time.




Ben,
ReplyDeleteDo you think it would be a good idea to increase the 'Max. Equal Modification' parameter of the SequestHT Node? (5, 10, 20???). Assuming there could be quite a few peptides with multiple 15N(x) mods per peptide, I guess this could significantly affect the results. I'm not sure whether or how the FDR would also be affected, as the search space will also change. Maybe I (you/we) should try it on that Pride dataset to compare the results. Your thoughts???
Cheers
Alex
Alex,
DeleteIts real tough to say. Honestly, if you gave it a try and it helped/hurt, I'd love to know. Ultimately if your max delta CN is still at 0.05 you are still only going to squeeze out one or two (at most) of the best SequstHT searches for the FDR calculator, so the hope is that we'd just get the best match and FDR wouldn't be affected. The reality? Honestly, I don't know. I keep planning to open the max delta CN up to see if it hurts/helps results for multiple PTM studies but I haven't had a chance to do it. Sorry if this isn't real useful...
Maybe I'm confused, could you explain (better than the PD Help) what you understand by the 'Max. Equal Modification' parameter of the Dynamic Modificatgions input in SequestHT Node? I assume that a value of 3 (preset value) would not allow, say, more than three 15N(1) mods (on A, D, E ect) per peptide.
ReplyDeletehope we can follow up on this!
Alex
Alejandro,
DeleteYes, I think you are right and you have a very important point. If you have maximum equal dynamic mods of 3 then you can only have 3 modifications for a single peptide. (ptmRS seems to be able to bend this rule, but that is a completely different point), then you will run into a problem rapidly with the dynamic mod on every amino acid. The workflow that I've gotten to work assumed 100% integration and I used these modifications as static. A customer of mine I worked with recently did something similar to my approach (I don't have complete details, this isn't a technology I've ever used personally) but if you wanted it to work as dynamic I think you would need to turn up the number of dynamic mods. Hey, I think I have your email address from a previous conversation, but why don't we take this offline? Shoot me an email at: orsburn@vt.edu. I'm traveling like crazy right now for work but I might have some time next week where I could block off some time to talk about this.