Thursday, June 30, 2022
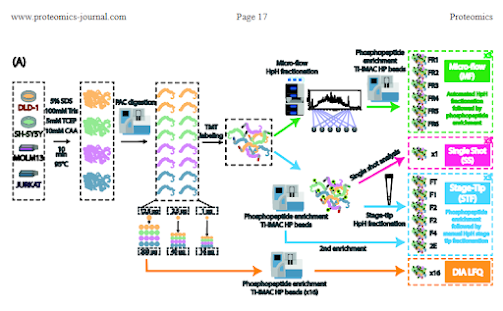
Optimal strategies for labeled multiplexed PHOSPHOproteomics!!
Wednesday, June 29, 2022
Using transcriptomics to REDUCE databases in proteogenomics!
This new study in Genome Biology is, on the surface, probably counter-intuitive. Our smallest databases are the nice reviewed ones from UniProt/SwissProt. When we start looking at databases that are a little (lot) more biologically relevant because of things like genetic variation, it is easy for the input database size to blow up to astronomical proportions. We frequently use an input database with millions of entries, which requires special class based FDR and a lot of computational power (2 million are known cancer-linked mutations, so they're just little snippets of sequences). When we start to toss in databases from those same samples that were derived from next gen DNA or RNA, things tend to blow up. BTW, neither is nearly as clean as you'd guess given the fact we're on "3rd gen" sequencing technology with 1TB of data coming off per sample with these new sequencers. There are fundamental questions being asked right now like -- wait -- is the genome way way way way more complex than we every thought, or is Illumina generating less and less relevant data with each generation and more literal garbage and hoping to cash out before someone stops to consider that the latter is the simpler explanation. Today's data density might give them another 10 years because it will take that long to process the data from 4 patients.
This wasn't supposed to be a genomics rant, but while I'm going -- long read sequencing is the way to go for us, y'all. Illumina and whatever the thing is that Thermo sells that no one uses, generate really short read sequences. 6-frame translate those little things (reducing them /3 to get amino acids) gives a lot of tiny annoying things to search against. PacBio and NanoPore both produce much longer outputs and it is transformative for us, both by reducing a ton of redundancy and giving us more sequences to match against.
All of the words starting with the 2nd sentence were meant to impart the fact that, unless I've been doing it totally wrong for 10 years, which is completely possible, proteogenomics databases shouldn't get smaller. They just keep getting bigger. It would be awesome if there was some way, any way, to reduce them.
There is a lot here, and the paper tackles two different concepts. The first is a recently proposed strategy for database reduction that I won't go into because they don't like it. The second big concept they use -- is one where they utilize the transcriptomics (RNA) to reduce databases. The logic is that if there are no transcripts expressed for that gene, it seems silly to go looking for that protein. Using this approach they find a "more sensitive" peptide detection rate (you'll see their terminology and it makes sense shortly into it) even using standard target decoy based approaches.
Big caveat here, of course, that if you are perturbing a system by, for example, irradiating cells that would induce a rapid response that would shut off transcription you definitely shouldn't do this. Proteins with long half-lifes relative to their oligo counterparts would still be hanging around, and then you wouldn't have entries for them. This is just the first example off the top of my head for what a bummer thinking about these systems in a biological concept is probably like.
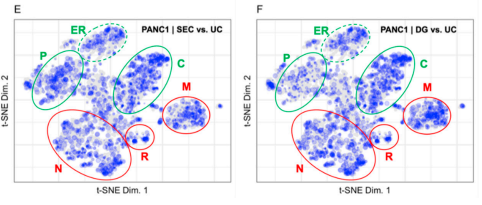
Also, I am not sure what the figure I chose for this blogpost is displaying, I really liked their choice of colors.
Tuesday, June 28, 2022
HUPO2022 -- Quintana Roo deadline next week!
Next week is the abstract deadline for HUPO 2022, which is being held in the Mexican state of Quintana Roo. This remote and obscure location was chosen as it would not flag the attention of managers and administrators who might question your underlying motivation for where you disseminate your newest research in Human Proteomics. We can't go to all of the meetings and we know that we only think about which meetings best aid in the advancement of our professional goals and our field before considering any other factors. While we all know this, these simple facts may be difficult for others who don't understand the level of devotion we have to our craft. They might, for example, question our choices between two very similar sounding conferences with similar sounding goals and names if we chose the one in a famous beach town in December over the one in Chicago in February.
Therefore, when time came around for Human Proteomics to be in the Americas again, the powers that be saved us any questions by choosing a town in largely unknown state of Quintana Roo in Mexico to make sure our motivations are not questioned. We can now drop right into a quiet and secluded location with a surprisingly convenient airport where there will be no distractions and just 100% focused Human Proteomics. Bravo, HUPO board, bravo!
Deadline is next week! (7/8)!!
Monday, June 27, 2022
ACS Special Issue Deadlines coming up!
Probably just leaving this here for me so I don't forget these coming up:
Software Tools and Resources (round 3!) has a deadline of September 30, 2022!
Sunday, June 26, 2022
Multiplex 29 proteomics samples by using every reagent!
Around my ever increasing horror watching my country, the one with the most nuclear weapons, fall every day to the demands of a minority of people who live their lives by literal interpretations of very small excerpts of an ancient and poorly translated book that they clearly have not read, it is hard to focus on science, but for my sanity I'm going to try anyway.
When I saw this new title I was SUPER EXCITED....
It is tough to create new multiplexed reagents, while more than the ones we commonly talk about probably exist the ones that we talk about are protected by some extremely observant lawyers. As I might have mentioned before, I've experienced the keen observation skills of said lawyers first-hand and what I type in these boxes has to, at some level, be guided by considerations of what might end up making them get all jumpy and litigationy again.
The number "29" probably should have tipped me off prior to the abstract that something fishy might be going on here. We've got 11-plex and 18-plex reagents now. Which to use?
At first you might say, as I did, f******************k thaaaaaaaaaaat. How would you process the data? Why would you ever double your reporter background interference from coisolated peptides? 126-131n literally just doubled.
Okay, but somehow the data looks good here. Maybe this is worth reading more of? They probably used real time search based MS3 or super tight ion mobility? Nope, they used a Q Exactive HF with 1.0 Da isolation windows an the JUMP masstag software.
The authors used a two proteome digest to pressure test their quan with E.coli labeled with one multiplex reagent and human brain labeled with the others and -- somehow -- the quan doesn't look bad. I don't think that with my software of choice that I can replicate this. I'd need to have my 4 tags that are usually static set as dynamic which would do wonders for my search space and FDR and it would take a lot of post-processing to get this sorted out. I might be wrong, though! And -- if you're in a pinch, maybe this is something to keep in the back of your mind?
Monday, June 20, 2022
Patterns in proximity labeling (and other experiments) improve when overlaying on LOPIT data!
Yo. This paper is super dense and it took me a little while, but I think this is a positive concept, particularly for y'all out there APEXing and BioID'ing stuff.
I might be too old for using nonlinear dimensionality reduction in my daily life but these smart young people with all their neuroplasticity are getting the hang of it. I think I've posted this before, but here is a good video introduction to these concepts. (All of this is just an aside, but it is really cool seeing t-SNE's in proteomics that make sense :)
Check this out, though. You know all the data people have been acquiring using LOPIT (localization of proteins by their organelles, or something, I'm at least close)? These data aren't just a neat trick like I thought. This group reuses LOPIT data to better understand other data! Think about where most of our annotations come from that say "this protein is in the cytoplasm, this protein is in the nucleus, etc.," I'd bet you $4 that it didn't come from proteomics data. It is more likely that in 1954 someone tagged a yeast protein with uranium and before they died at their microscope, they were pretty sure that protein was at the yeast membrane. Then someone in the 90s when all crazy with BLAST on their NetScape browser and found a 10% sequence match and 1e-74 whatever crazy metric-less score on BLAST that makes everything sound more significant than any metric ever and now that human protein is annotated as membrane forever. We should probably think about updating some of that stuff with LOPIT data....
When you do? These authors demonstrate multiple examples where evaluating quantitative data in this subcellular context further strengthens the conclusions of earlier studies. Even more interesting? There is a strong example where the conclusions you can draw from the data seem very different when thinking of the protein in an organellar context rather than as a list of ups and downs.
The trick here is how to apply these maps without having to bother with updating my R packages which I'm sure are out of sync between PCs by now....
Sunday, June 19, 2022
Find the genomic variants that make it to peptides -- with MaxQuant!
Step by step instructions for how to do something -- in MaxQuant?!? That's great!
Genomic data has just as many stupid formats as we do, but their files are often bigger. I've built a couple of proteomics studies from crappy sequencing data on repositories and I can't remember what the difference between a .fasq or a .bum is without looking them up. Chances are I'll just download the wrong one and will realize it is wrong when I can't convert it.It's all here!
Saturday, June 18, 2022
Perspectives on de novo antibody sequencing!
A lot of antibodies and antibody drug conjugates and whatever they call those other things coming your way? Thinking about how best to analyze these frustratingly impure things? It is crazy how a fine tuned evolutionary mechanism optimized over billions of years to make variable proteins to combat a near infinite number of biological obstacles can end up making varied proteins despite our 40 years of effort to make them make one thing at a time.
However, utilizing these things for our own designs is awesome and if you start accepting antibody characterization projects it can open the flood gates to crazy proteins.
Where are we at on de novo analysis of endogenous antibodies? Here is a great perspective from a team that knows a thing or two about it!
Friday, June 17, 2022
High pH fractionation of 6 microgram of peptides on 96 well plates!
I've been looking absolutely everywhere for something like this and I think I've got everything I need already sitting here to pull this off! Paper link!
On closer look this isn't exactly what I was looking for, but I think that I can use this as inspiration because their goal is different than mine and they demonstrate how remarkably flexible C-18 and HPH fractionation can be. They essentially elute their peptides off of immobilized C-18 and pull the supernatant as the fractionated. Super cool, right?!? Sounds a little complex, but they boost their peptide coverage of interest from low sample volumes, so there are applications for this kind of fractionation for sure.
Thursday, June 16, 2022
Mouse proteome draft map!
Whoa! That's a lot of samples! And they did phosphopeptide enrichment of all of them?
Okay, so if mouse proteomics isn't your thing but cancer is, there is a second resource as well. Check out this web deployment of Pacific!
Wednesday, June 15, 2022
Is fancy trypsin the best science scam of all time?
When I was in high school I worked at a grocery store at night with a bunch of other poor redneck kids and we did typical stupid redneck kid things before and after work. My coworkers had the biggest subwoofers that would fit in their Ford Festivas or Chevy Cavaliers and we'd test to see if you could hear the bass from these in places like the walk in freezers inside the grocery store. Being a nonconformist, I spent all my money on gasoline and new rear tires for my idiotic 79 mustang that had way too large of an engine for the 4 spd transmission that was intended for the stock 2.3L shared with the Ford Pinto. Why did that have a shared bolt pattern with the pre-emissions small blocks? Who knows?
My friends spent a lot of their money on things like gold plated amp terminals. Or gold plated wire and I always assumed those were real things until James Randy went on a crusade about it and proved humans can't discern the differences (if they even exist), big shoutout to Dave for tipping me off to this years ago.
Why would I ramble about this scam at the end of my blog hiatus? Because there might be a parallel. As some in proteomics are shedding off the (largely unnecessary?) burdens of nanoLC there is a downside in that you need to load more peptides and may need tons more of that expensive trypsin. Even if you aren't scaling up sample sizes, we're being pushed to higher n and trypsin prices aren't going down. Last year at ASMS2021 I got to see some great analytical flow data at a poster by Matt Foster and he tipped me off to the fact he's using "lower grade" trypsin and his data looks sick.
I got some cool waste material from our group and set to work with trypsin that cost me $40 a gram. And, you know what? I can't tell a difference AT ALL.
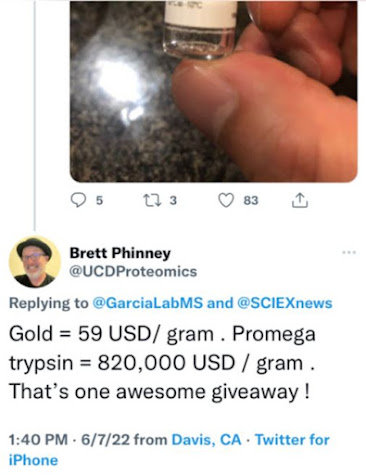
This is probably my worst run so far. I think it is from heart (randomized tissue samples) but given the proteins that would be my guess just looking at it.Are we all being scammed? At ASMS2022 they were giving out some free new super duper trypsin samples. Brett, one of the few mass spectrometrists to not get COVID at ASMS because he got it right before, did this math.
That's kind of a lot of money.
Sunday, June 12, 2022
Trap column intact protein analysis!
Okay y'all, get this. Those trap columns that you just use for making sure your collaborator's samples don't ruin your nice instrument are actually just normal columns that are smaller! Not kidding. You probably wouldn't have realized this from the fact that they say misleading things on them like what resin is inside them and length that they are.
This royal chemistry society advance exploits this fact!
In this advance, this team skips the desalting steps for intact protein analysis and -- get this -- they run their protein over a column while allowing the first part of the liquid that goes through the column TO GO TO WASTE! The salts from their intact protein don't go into the instrument. It goes to some waste thing. Then they elute off of that trap column as if it's a short little column. And the proteins come off of that column and into the mass spec saving untold amounts of time.
I largely like this paper because I think that it you could potentially read my impression of it in two different ways. 1) If you didn't know that you could do this, legit, bravo to these researchers and this nice data. Not kidding. Not being sarcastic. I will be citing this paper all the time going forward.
2) If you've been doing intact protein analysis like this for the last 10 or 20 years and are thinking -- obviously?? OMG, WTactualF?!?!? this should impress upon you how poor we are as a field of passing our information on in a structured and universal matter.
Mass spectrometry is still more like an art than a science in a lot of ways and that isn't a good thing at all. Maybe there is a paper that shows this would totally work out there? I can't think of one and I couldn't find a paper with a Scholar search this morning.
I'm absolutely going to cite the shit out this paper.
Thursday, June 9, 2022
Agilent ASMS2022 hardware releases -- new QQQ and GCMS!
GCMS is something that I think most people in proteomics are probably surprised people still do, but it absolutely has places in chemistry. Wondering about that lama oil you bought from the old man with pigtails at the farmer's market that you largely purchased just so he would stop standing so close to you that kind of smells like acetone? Easy to figure out that -- YUP. That's a lot of acetone! Try doing that will LCMS!
Next question: WTF do they feed that lama? Aaaaand....which government agency do you even contact to explain this situation to without sounding crazier than the close talking pigtail grandpa?
Despite the clear importance of GCMS to things like lama protection and the petroleum industry (or CBD extraction from hemp which is sometimes performed by people who make lama oil people seem well grounded in reality and scientific capabilities) there hasn't been a lot of development in this space, besides in the ultra high end market (GC-WTOFs and GC-Orbis). It's cool to see that Agilent is pushing forward into this space with the release of multiple new GCMS systems.
In addition, a new small, but high end performance QQQ dropped at ASMS. There is more info on these releases here if this is your thing.
Wednesday, June 8, 2022
ASMS Big hardware release #3 -- ZenoTrapping SWATH
My coverage/guest blogger coverage of the COVID superspreader event otherwise known as ASMS2022 was impacted by some craziness in my schedule and the fact that no one I know has felt very well recently for some reason. I'll do some backtracking as I find time.
On the hardware front, one move that likely only will surprise you because you probably assumed a ZenoTrap on a SCIEX QTOF would be able to do ZenoTrapping + SWATH, SCIEX rolled out ZenoSwath. The one in our lab (proof it is here in the only mass spec lab in the world with magic pink carpeting [magic because it keeps the asbestos in the floor and out of our lungs!] can't yet ZenoSWATH, so we are puuuuuuuuuuuumped for that upgrade!)
The ZenoTrap is inline after the quad and after the collision cell and right before the TOF. The physics of the ZenoTrap is really cool because it essentially slows the ions into accumulation rather than pushing them to a dead stop with a hard gate. This is super important for my stuff because it compensates for the small time of flight effect of fragment ions of different sizes, allowing the ZenoTrap to eject both high and low mass fragment ions into the TOF in one go. Somewhat less important for most SWATH experiments, but a feature that helps me out a lot.
It looks like from the online info that you do necessarily have to lose some speed when using ZenoSWATH vs regular SWATH, but 133 Hz isn't all that bad for a 10x boost in low abundance signal.
This is an aside because I just got data from this. I'll be honest, while EAD and the E-I-E-I-O fragmentation stuff sounded like a neat trick (giant magnets force a charge onto your peptide and you get ETD/ECD fragmentation -- you can actually use your phone to detect the magnets, they're that powerful) it wasn't why I wanted a 7600. I need higher intrascan linear dynamic range. I had some open time on Sunday night and reran some leftover samples with EAD and....it's probably the best looking ETD-type spectra I've ever generated.
This is following using a non-fragment filter and exclusively plotting c/z ions. I'm not picking and choosing, they ALL look like this. I don't have any good PTM data, I just used it for regular old peptide ID. I'll drop some comparisons that make sense later!
Tuesday, June 7, 2022
Guest blogger on the ground at ASMS2022 -- Software updates!
First off, let’s start on the software side.
Fragpipe 18.0 Just dropped from Alexey, bringing a host of new features with it. There is now integrated spectrum visualization of results, a “Headless Mode” that enables fragpipe to be run without the GUI and in the command line so now you can spin it up on your server, and something called diagnostic feature discovery apparently giving you a boost in IDs. New verison can be found here (https://github.com/Nesvilab/FragPipe/releases)
Ben interrupting: There is currently a search for mnemonic devices to assist with the spelling of Alexey's last name. Word is he doesn't even know how many "i"s are in it. (As someone with a silent "r" in my last name, I'm allowed to say things like this, the rest of you should try being less insensitive).
Ben interrupting: For my impression of CPU based machine learning rescoring with Impetus in PD 2.5, please see this post. Y'all, more peptide numbers is cool and all but not if we can't back them up. I am pumped for PD 3.0, but more numbers without evidence is just going to burn this whole proteomics party down right after we got credibility again. Sorry to be a jerk again. I should really check it out again.
The Kuster lab is putting out something that may change the way that we do TMT and label free analyses....Instead of doing a match between runs approach, heavily depending on chromatography, they developed a spectra clustering algo called SIMSI-Transfer. So from my understanding, the spectal scan are grouped into clusters by their similarity. If a spectra doesn’t have a match in the cluster from a search, the identify of that spectra is assigned based on which cluster it belongs too. They showed a >35% improvement in PSMs, >15% improvement in Peptide IDs and >5% protein IDs over match-between runs! This is available right now on github https://github.com/kusterlab/simsi-transfer !
Finally, I have no idea that Metamorpheous could do this, but did you know you can search raw files that were collected in a top down method and a bottom up method at the same time and it kicks ass at identifying more proteoforms than a single search of top down run???? Basically you add your bottom up and top down data to a search but change the protease to topdown for your top down file and then let it fly! They are reporting that a Top down search along identified about 121 different proteoforms, but when you added the bottom up, BAM! Over 8,000 proteoforms identified!
Cool advancement on the software side!
ASMS 2022 Release -- TIMSTOF HT
There are a couple of ASMS hardware launches this year that I'm not finding quite as simple to understand. Something like "this is 20% faster" is pretty straight forward. Something like "we're allowing you to see a new math from the inside of this instrument now that every instrument we've manufactured for the last 18 years has generated, but we hid it, so now you should pay us a lot for us to unhide it because someone pointed out that charge detection is actually really cool" is a little less easy to understand. Oh. Actually, now that I typed it out I actually do understand that one better.
Changing topic! Let's talk about 114 out of 117 of the acronyms on the Free Dictionary for the letter's HT and let's try to guess what it stands for on the new TIMSTOF based on what it is!
The TIMSTOF HT has a new (4th generation) TIMS cartridge. The vendor says higher dynamic range than generation 3.
It has a higher frequency digitizer which might be really super cool. In my head, I think that the limitation in resolution on TOFs is often how fast they are able to distinguish between an ion that gets there in 81 microseconds from one that gets there at 81.1 microseconds, so you need faster digitizers to get there. However, the best I can tell from the outside we haven't heard about resolution increases. What we've heard about is loading micrograms of peptides on a TIMSTOF. On the Flex, we aim for 200 nanogram when using nanoflow rates. I think for some long gradient stuff we've went as high as 400 nanograms, but anything above that hurts us.
If maybe the TIMS is suited for high tension or heavy timber?
You really have to sort through a lot of these on Google images to find one that isn't cringy.
Awesome! Okay, I bet it's this, though.
Micrograms of load seems to better allow microliters of flow, which is what we need to get proteomics onto the ground of routine labs, which I'm excited to see is finally a hot topic.
Microliters of flow also allows us to typically get to lots more samples per day which will largely reduce cases of hypertension so common in vitamin D deficient mass spectrometrists.
Using higher flow rates allows us to spend less time tinkering with pesky nanoLC systems which would allow us to sleep better, therefore increasing our hydroxytryptamine (serotonin) levels? That's pretty deep, maybe not that one? I'll keep thinking about it on my commute.
Monday, June 6, 2022
ASMS 2022 - Is Waters is joining the proteomics party again finally?
The last few years have been a lot of fun for mass spectrometrists because, all of a sudden, proteomics isn't a one player field. Shocking advances from Bruker, that most of us didn't even know were in business and the out-of-the-blue drop of the SCIEX trapping ZenoTOF have made every proteomics purchase something you actually have to think about.
Is this the year that Waters joins the party? It might be.
Those 3 little letters signal something really exciting. Last year Waters dropped the DESI powered W-TOF for mass spec imaging at up to 240,000 mass resolution. Not the fastest thing ever, but still enough that a lab 2 miles from me in Baltimore got one and already had data to present this Sunday.
I haven't seen a ton of data yet, and it has largely been from their Australian marketing people. What I did see, however, was resolved fine isotope structure -- on a TOF. And mapping of a tryptic digest of the NIST mAB standard using 1ppm mass tolerance windows.
Resolution on the W-TOF is gained by performing additional passes, so it goes faster at lower resolution. I don't have time for specs, but it might be worth taking a look at. They also launched a MALDI for this.
AND -- a G3 system. We had a G2 system in my lab in 2011, and while they've tweaked the Q-TOF over the yeras it has always been a G2. What'a a G3?
It looks like a Tweaked G2 with a new IMS system and a really interesting charge state controller for automatic adjustment of the sensitivity (through scan averaging? that's what I'd guess, but I'm not sure.)
I gotta run, but details of other cool hardware drops are in my "to read" folder.
Last thought, though, and really the inspiration behind this post this morning: If it's time for you to upgrade your instrumentation, and you really have to evaluate hardware from 4 different vendors -- like, for real, look at specs, because they can all do what you want -- when does this competition lead to competitions for pricing for us?
It doesn't seem to have happened yet, but it has to, right?
Sunday, June 5, 2022
ASMS Guest Blog post for Day 1 (Saturday Machine Learning Course!)
I had to cancel ASMS pretty much last minute this year for family things, making it the first year in a lot of years that I haven't been there. My photo storage app keeps popping up cool stuff every day to remind me of how much fun I've had at ASMS over the years with great people that I can't wait to see soon.
When I realized it was really going to be a no go for me, I put out a SOS to one of my very favorite people who is on-site in Mosquitonappolis for the whole thing to keep me and the blog in the loop.
This was his first ASMS poster and abstract during his M.S. a couple of years ago, a photo that popped into my "this day over the years" thing this weekend.
@SpecInformatics will be providing insight into ASMS from the ground to keep me, and this blog in the loop:
Guest blog post day 1:
ASMS 2022: Machine Learning Short Course
Happy 2022 ASMS y’all! It’s the most wonderful time of the year again (even though the last one was only 8 months ago) and I am pumped to see everyone.
Machine learning has been a hot topic that our community is finally fully embracing as an important tool and not just a buzzword that people throw into papers in an attempt to get into a higher impact journal. Many people are familiar with percolator, prosit, and DIA-NN and how big of a contribution to the field they are but what is really going on under the hood? Going through the respective publications, there is a ton of computer science jargon that I expect the average mass spectrometrist can interpret as a foreign language. Now if you're like me and have dipped a toe into this realm it sometimes seems that what you get out of using these tools seems like some magical space wizard took your data and generated some info and figures that will hopefully tell you a really cool story. Want to know more about the spells and incantations that Machine Learning makes on your data? Well this new short course at ASMS 2022 is one you should take!
This is the first year that ASMS is offering a 2 day Machine Learning short course taught by Wout Bittremieux and Will Fondrie and you definitely can’t tell that this is the first time they are teaching this course because it has been on-point. The first part of the class was basically a proverbial Rosetta stone that explained the difference between supervised and unsupervised machine learning, the flexibility of models and how it correlated to bias and variance, how to properly train and test models and what the heck a loss function is. We then moved into Dimensionality reduction. Many of us use PCA plots with colors and circles and say “See! My samples are different!” Well, Are you using them appropriately? When do you want to use other components? Wait, what is a PcoA or t-SNE? Why am I not using them?? This course touched on them all and I definitely need to rethink how I have used them and how I will use them going forward. Day 1 was mainly going over the theory behind each of these methods,the advantages and pitfalls of each and how to appropriately train and test your models. Day 2 is when we really got into it using logistic regressions, support vector machines, random forests and neural networks. Well if that wasn’t exciting enough, Wout and Will made some awesome tutorials online for us to test out and if you are curious yourself, they allowed me to link it here: (https://wfondrie.github.io/2022_asms-ml-short-course/)! We even got a chance to test out if we can train a random forest model on the 2020 ALS challenge that Ben put on. I have been to I think 5 short courses held by ASMS now and I think this is one of the best I have been too and I encourage anyone from any level to attend even just to generate ideas that you can send over to a bioinfomagician and understand the data that you get back. Great start to ASMS and ready for the rest to start!
End guest blog post
Saturday, June 4, 2022
The (first annual?) Johns Hopkins Mass Spec Day!
ARE THE WINDS CHANGING? Bob Cole set up a mass spec day and PEOPLE SHOWED UP!
These things have been tried here and then over the years in town and it's often been Dave Colquoun(SP?) and myself in the audience and maybe 3 grad students who look uncomfortable, as if they accidentally went to the wrong room and feel bad leaving something so poorly attended.
Over 100 people watched remotely and a decent sized room was filled. Some of the highlights from my perspective were:
Peggi Angel showing more of her group's amazing heterogeneity data (proteins in context, but this time from the imaging perspective).
Ryan Kelly (stranded in Belgium due to this stupid virus) and Peter Nemes demonstrating more of their push toward understanding systems at the single cell level.
Marian Kalocsay showed the most impressive use of APEX labeling technologies that I've ever heard of. I'm not 100% how much I can talk about here because he was clear a lot had not been published. His group is pushing our understanding of how biological systems work -- in almost real time -- by being very very clever about how, where, and when they apply APEX. Definitely watch out.
Brian Foster described targeting in a "post proteomic map" world and how to leverage tools like Panorama Web, PASSEL, the SRM Atlas, and SRM and SureQuant(TM,R), etc., to get to the protein information that you want.
There were other great speakers and I think someone called out sick because I got a chance to speed ramble through how we're using single cell proteomics to figure out drug mechanisms that we've been stuck on.
AND I learned a total life hack. Someone left the door to the sound booth open the whole day and I just hung out in there. The chair was way more comfortable, it was perfectly in the middle of the screen, where everyone else was to the left or right of a main central set of stairs and I had a full desk for my laptop and notes.
Friday, June 3, 2022
Double barrel 10uL/min TIMSTOF DIA -- 4k proteins in 15min!
Yikes. I might have to find something else to do because Proteomics seems to be growing up. CVs, data completeness, and instrument utilization (IU) measurements?
IU is the actual amount of time that your instrument is collecting useful information. If you've got an EasyNLC it doesn't count that 15min to 7 hours that the pump is kind of thinking about whether it actually is at 0 bar before finally starting to flow solvent again.
This group is getting 90% IU and phenomenal results by using 10uL/min flow rates and an extremely well defined double barrel chromatography setup. Totally worth checking out!
Thursday, June 2, 2022
Convert ZenoTOF data to centroided MGFs for you can process with anything!
Do you have one of these screaming fast self calibrating EIEIO thingies around and are having trouble processing the data?
I think I have it sorted out so that the data from Proteome Discoverer, MetaMorpheus and FragPipe closely match what we're getting from the OneOmics Cloud for each file. Of course, this just makes an MGF so you lose the MS1 level data, but if you are doing spectral count things or TMT/iTRAQ then you don't care about that.
Here are the tricks for working with these data. I like the .wiff2 files because they put the charge state in the header where I expect it to be (every run outputs both wiff (to not break everything for historic software, I think) and .wiff2.).
1. You want to convert in the same output folder where your .wiffscan data is (the .timescan files might also be important, I dunno). If you transfer files, transfer everything. Don't just move the .wiff2, it won't do anything.
3. I'm using the MSConvert PeakPicking with a really tight tolerance. I don't think this is necessary for everyone, but I was trying to see if I tuned out the TOF whether I could resolve the TMTn/TMTc ions. Your files will be smaller, I think, if you raise this a little, but this works great for my stuff.
7. Referencing #1 above. When I submit data to MASSIVE I think I'm going to zip my .wiff2, .wiffSCAN and .timescan for each run together and upload those like that to keep them organized.
To load this into MSConvert, go to your File search menu in Windows and find the seemingly random place the MSConvert ended up and place this .cmdline file in the same place as the other ones, then open or close/reopen MSConvert. You'll now have a default pulldown for 7600 file conversion.
As always, zero guarantees that this will work.