I suspect that we're about to see the floodgates open wide now that SomaScan and O-link have gotten some level of acceptance for their ability to do huge throughput studies and here is a taste of what we can expect!
What did they do? Well, they somehow got 300 or so participant samples of 3 types (blood, plasma, and urine) and they did all the -omics! Then they built this Shiny app!
Before you do anything else, I STRONGLY suggest you make a network and then click on the center thing in that network and drag it around.
I wish I had time to make a .gif of it! Here I've clicked on the star and then just hold your mouse clicker thing and drag it around the screen really fast. I don't know how they did, that and I know someone to ask. It is really really fun and, unless I'm missing something big in this preprint, supplemental data or clicking around in the app, it might be all that they did with this patient cohort.
I swear, every one of these next gen proteomics papers that I've really put any time in makes me feel like almost 30 years of headbanging finally did catch up to me the way my Mom said it would. I get to the end and I can't figure out why they did it, what they were trying to accomplish and how it could possibly benefit anyone or anything in any possible way.
Again. Probably just mushed my brain, but I did enjoy this cool Shiny web effect and I'm using it later for sure!
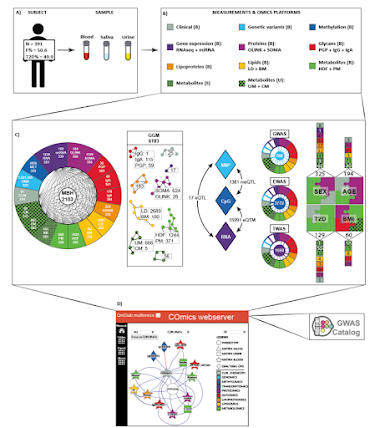


Dragging the items around the screen is the most fun
ReplyDeleteThank you for your interest in our paper. Let me answer a few of your points:
ReplyDelete"Here I've clicked on the star and then just hold your mouse clicker thing and drag it around the screen really fast."
That's done using the R package VisNet under Shiny - the full code of our server is on GitHub: https://github.com/karstensuhre/comics
"A moderately sized study that was obviously really expensive and designed by a committee with some really great intentions and some solid bioinformatics visualization people!"
LOL ... a committee???? Far from that ... we ran a pilot study in 2012 to just do metabolomics in urine, saliva and blood, but wisely collected many aliquots for future analyses. Then over the next ten years I could send samples to any new platform that came to the market.
Weill Cornell Medicine in Qatar had the first Somalogic platform outside the US, and also ran the first Olink platform in Middle East. Our own genomics core ran genotyping and epigenetic arrays and performed RNAseq. In some cases companies even ran samples for free, so that we could compare them to others.
"I get to the end and I can't figure out why they did it, what they were trying to accomplish and how it could possibly benefit anyone or anything in any possible way."
Read the full paper - it's all there. Briefly - we show how all these multiomics platforms compare to / complement each other.
Check out our homepage:
http://www.metabolomix.com/comics/
It has a number of work-through use cases.
Thanks again for your blog,
Karsten.