Shoutout to @MatthewCollins@mastodon.social for his post on this crazy awseome thing!
Do you want to just 3D model any protein you feel like? It has never been easier than this thing that is powered by Meta!
Not only can you look up your specific protein, you can essentially blast a FASTA sequence against their database and then 3D model that.
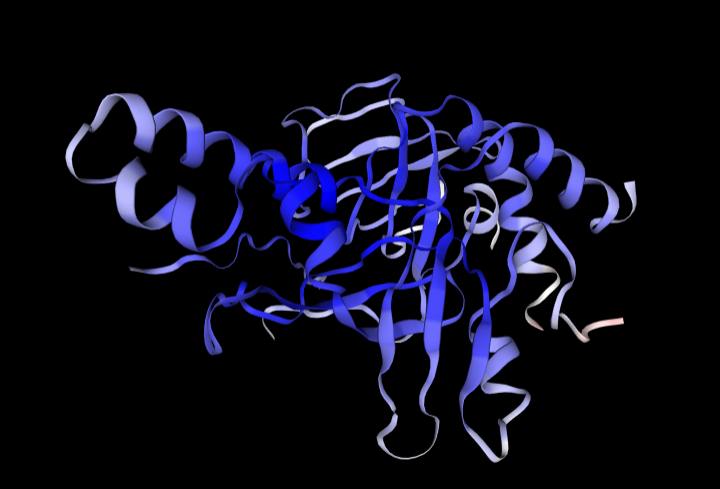
Check out what happens when I model an essential DNA damage protein I can't seem to get very good coverage of and I don't know why (possibly PTMs everywhere, and maybe hyperphosphorylated, but I don't know yet).
Now, we can go to SWISS-Model and pull a couple of Cryo-EM based ones!
It looks bad at first, but if you ignore the first 51 amino acids you can see it actually did a really good job. The three main Rotinis line up reasonably well, though there is a difference in whether they are linked by Macaroni or Mafaldine folds. The cryoEM that I pulled had a short ligand in it that appears to interact with the wobbly Trofie terminus predicted by Meta which may be what stabilized it into the Tagliatelle structure.
YOU KNOW YOU WANT TO MODEL THINGS IN IT! GET GOING!

No comments:
Post a Comment