(From this original article in the Guardian)
....okay...well, glyphosate is a ridiculous molecule to analyze. I've never gotten it to retain on any chromatography at all. The only way I've ever gotten anything close to reasonable signal on it was by analyzing it in the flow through and using a really great catch all mixed-phase chromatography to retain just about everything else.
What are the odds that the group who did this analysis are just quantifying the wrong molecule?
Here is the report. Hopefully they don't know what they're doing!
It is worth noting that the carcinogenic aspects are still somewhat debated, but there have been a flurry of successful civil suits in the last few years from farm workers who have been exposed to a lot and ended up with very similar cancers.
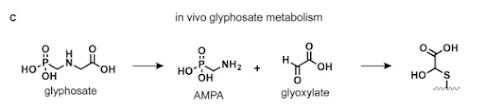
At a functional level, this paper from a couple years ago provides a ton of insight.
They also broke out stable isotope glyphosate from Cambridge Isotopes and injected it into mice. They tracked the small molecules and they did a lot of proteomics using a variant of MuDPiT on a QE Plus system and did both lipidomics and proteomics on top of targeted analysis (it is a really nice study all around).
What they find is a pretty clear mechanism of action....
(that craziness on the far right is stuck to cysteines....)
And these effects, while they can't be ruled out elsewhere, appear to be the most pronounced in the liver and to set of important detoxifying proteins, which probably would better without some craziness stuck to all of their cysteines....
On the positive side, if you are doing human proteomics on Americans, you know a new PTM that you can be looking for, particularly in liver samples, though reduction/alkylation might strip that mod off? I'm not sure and it's already 5am and I've spent way too much too much time on this rabbithole.





No comments:
Post a Comment