Tuesday, July 5, 2016
Tracking cancer mutation status in historic proteomic datasets!
(Image stolen from this paper)
p53 (or TP53) is a central regulator of just about everything. It is also mutated like crazy in cancer. There are regions of p53 that are so-called "hot spots" and are much more likely to be modified than others.
A super good review of the protein is this one from Bernard Leroy et al., (it is a couple of years old now). Why am I rambling about this when it isn't new? Honestly, I had a conversation the other day and it centered on how much of a cancer cell line's mutational status could be forcibly extracted from historic proteomics datasets.
Wait. I'll back up. In my extremely basic understanding of how cancer works -- A normal cell is going along just fine and it randomly picks up some DNA damage here and there. Normal consequence of life, metabolism and making new DNA. Typically the cell can use the simple doublechecking proteins and make fixes. If it is a big mistake the cell-cycle checkpoint proteins kick in and stop cell division while the damage is repaired. If it can't be repaired, then the cell is destroyed.
Stuff goes crazy when your checkpoint gene/proteins get messed up. p53 is intrinsic in this whole checkpoint thing. p53 gets messed up...and....now DNA damage that is picked up is copied into the next cell and you start seeing mutations all over the place! The weird part here is that p53 looks pretty easy to mutate.
I made a quick summary from the Leroy paper and from COSMIC
These are cell lines from the NCI-59 panel that I selected almost at random (honestly, cause they were the first ones that finished downloading a few weeks ago). p53 is messed up in all of these lines except for MCF-7 and H460. In all the ones at the bottom, they have the same stupid mutation!
That site in yellow is an R instead of an H.
Enough background and back to the question!
Question: Can I find out what the mutational status of p53 (and other important proteins, of course!) in a normal shotgun proteomics dataset?
Answer? Hell yeah I can!! -- with a couple caveats.
Proof? Okay, so let's start out by going to the NCI-59 cell line repository here. (Quick shoutout to Drs. Kuster, Meng, and Gholami for helping me out with some extra info on this fantastic resource!!!)
The original paper was in Cell a couple years ago here.
For the first run, I just took my normal Uniprot human database (that contained wild-type (WT) p53 and I added the R273H mutation. Cause I'm a resolution snob these days, I only went with the "Deep Proteomes" at first (these are Orbitrap Elite high-high datasets -- 24 fractions, I think, seriously nice dataset!)
This is what I get when I just run MCF-7 and U251 (MCF7 should be WT; U251 - mutant! See? Nice dataset (and I'm being lazy, this is just an old UniProt)
So...even in Uniprot, there are a lot of isoforms of this important protein, but only one peptide that corresponds uniquely to the R273H mutation. See the weird stuff, though!??! Where is the wild type?
This threw me for a loop. It isn't there. What? It has to have the WT protein, right? Yeah, it definitely has to be there.
Probably just something weird, right? So--- let's go to the other WT strain!
Son of a fish.....same thing!
Lets go to the XICs!!!
Well, at least I'm not craaaazy.
That is one peptide that should be carried over from the WT strain and any of the mutations. We'd expect to detect it...unless we don't want the WT protein around either!
Yeah! There shouldn't be any p53 around unless the cells are under stress (or the protein is mutated). If everything is cool, MdM2 should degrade it almost immediately. Link to this here.
Man, I read way too many cancer papers tonight working on this.
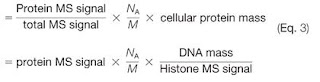
One last side note, though. Remember the Proteomics Ruler? If you can't find signal for any of the peptides from a protein -- at the MS1 XIC level ---
You're essentially dividing from zero. That is some loooow level stuff.
Wow. That was WAY too long.
1) Can you determine p53 mutational status? Heck yeah you can! And I lucked out, cause p53 mutants seem to lead to overexpression and then its really easy to find the mutational site!
2) With the caveat that there is a dynamic range to proteomics, still. (Hey! Anyone doing MCF-7 proteomics on a Lumos? I'd love to see if you can see these peptides!!)
Subscribe to:
Post Comments (Atom)








No comments:
Post a Comment