Wednesday, September 12, 2018
Rapid assessment of non-protein contaminants with Skyline!
Hopefully if you're doing proteomics you're always throwing in some great FASTA entries from cRAP or the MaxQuant contaminant database or have even generated your own list of stuff that you find in every water blank (or a combination of all 3).
Have you ever seen a way to keep track of the junk in your sample that isn't from sheep wool or gorilla keratin peptides?
Me either! Here you go!
Loads of reasons to read this paper.
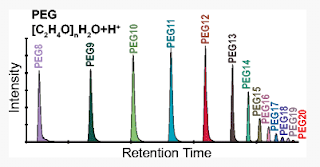
1) PEG is in just about every sample in some way. It's only when there is tons of it that it's a serious problem. This can help you keep track of this!
2) PEG is the first thing you might think of, but there are other contaminants as well. And this method doesn't just work for proteomics. It'll work for any LC-MS experiment.
3) The author totally pulls off a full (and awesome) application note as the single author. It's a great precedent for people with a bunch of stuff on their desktop that they felt funny about writing alone. Writing "I" a lot in a paper feels really weird while you're doing it, just because you're so used to reading "we". It doesn't come off as weird when you read someone else who wrote it that way.
4) In Excel you can =MROUND([Cell],5) to round to the nearest 5. Which no person ever in the history of the world has ever needed. You're welcome.
5)
Subscribe to:
Post Comments (Atom)

No comments:
Post a Comment