Woooo!! While most people are trying to get usable shotgun data out of their first actual single cell right now, there are a couple groups out there trying to do the impossible -- TOP DOWN proteomics from single cells.
During the Proteomics Show Road to Chicago we talked to both Alexander Ivanov and Ying Ge and I was surprised to find out they were both doing it -- and both have published now!
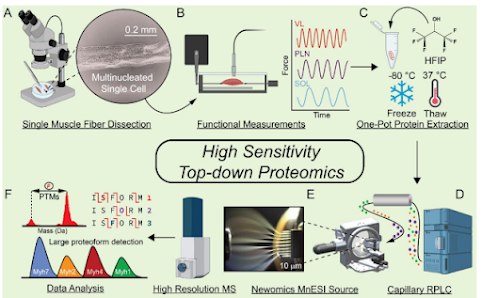
You can check out the new study from Ge lab here! There is an earlier preprinted version if you can't figure out how to log onto PNAS through your library....
Turns out you can get over the sensitivity hurdle for the device by using 8 different 10 micron ESI emitters. Also, the muscle cells appear to be sort of huge, so that probably helps.
However, they resolve proteoform level differences -- between different muscle fiber cells (!!!) in proteins in excess of 200kDa (!!!) I've only resolved a couple of proteins in my career above 200kDa and typically at the sort of resolution to say "yup, it's probably about 10kDa bigger than you said". What they've done here -- alone -- sets this study up in rare air. To do it in a physiologically relevant context? In single f'ing cells?
Huge props to this team. 100% recommended read that sort of resets some of the boundaries of what we can do today if we're willing to think outside the box.

No comments:
Post a Comment