This was sitting open on my desktop for a while and I kept skipping over it. You'd guess from the paper title that you would be looking at a secret magic instrument and a bunch of python doodads to process the data, right?
You aren't. This is a commercial instrument (Exactive UHMR) and commercial software (BioPharma Finder 3.something) used in a really clever way to work out an intact protein complex.
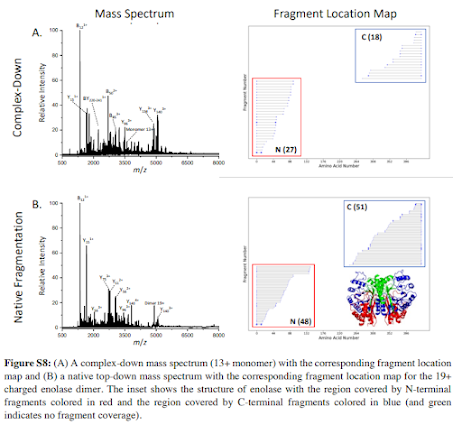
Where this really shines is how the authors work out the native monomer fragmentation patterns and then apply what they know to the tetramer. Check this out --
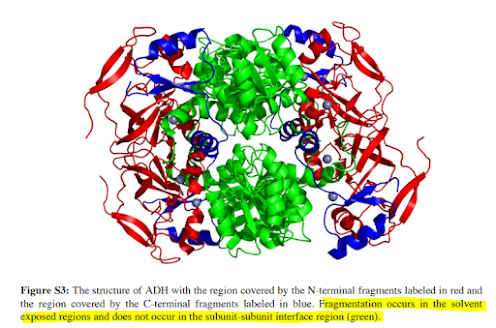
RIGHT?!? Makes sense, but I wouldn't have thought to check! Normally when you see really nice topdown work they used something that is 8kDa. This tetramer is ~150kDa! So here you are with the ability to study the intact monomer easily, the native monomer, get a solid high accuracy mass of the intact thing and see it's a tetramer. That's all normal stuff a lot of people do. But if you do the native complex fragmentation you can start to figure out how the darned thing is configured. What's internal in the complex? What is solvent facing? You can model this stuff with tools, chances are if you use 3 you'll get 4 different anwers -- and here you get direct evidence of which model is correct!

No comments:
Post a Comment