Wednesday, April 30, 2014
Abird -- little box that removes background ions
Apparently, people in Boston like to blow air across the fronts of their instruments. Unlike the little device in yesterday's post which blows away all of the ions that are produced during the crappy parts of your run, this little thing, the Abird, blows a slow supply of clean air into your source at all times.
And it looks like it helps! Y'all probably have known about this for a long time, but its new to me.
You can find out more about Abird at ESIsourceSolutions.com.
Tuesday, April 29, 2014
Want to keep your Orbitrap clean longer?
This is just awesome. Maybe other people are doing this, but I've never seen it.
Can you see what that is? It is a way of diverting the sweep gas so it is useful for nanospray applications! At the beginning and end of a run, alternative tune files are used at BRIMS. These tune files contain instructions to turn on the sweep gas. This appears to effectively blow away the stuff that comes off the column at the beginning and end of the runs and extends the amount of time they have between cleanings!
Want to set your own up?
It's the P-430 Union at Upchurch Scientific. This setup has been shown to work on both LTQ Orbitrap systems and the Q Exactive.
Correction (thanks guys!) its a P432, not P430. For more details see this new post.
Can you see what that is? It is a way of diverting the sweep gas so it is useful for nanospray applications! At the beginning and end of a run, alternative tune files are used at BRIMS. These tune files contain instructions to turn on the sweep gas. This appears to effectively blow away the stuff that comes off the column at the beginning and end of the runs and extends the amount of time they have between cleanings!
Want to set your own up?
It's the P-430 Union at Upchurch Scientific. This setup has been shown to work on both LTQ Orbitrap systems and the Q Exactive.
Correction (thanks guys!) its a P432, not P430. For more details see this new post.
Monday, April 28, 2014
qcML: an exchange format for quality control metrics from mass spectrometry experiments
More quality control software solutions! You know how I love this stuff! I'm about to board a plane to Cambridge for the PD 1.4 workshop so I'll admit I haven't read this all the way through, but what I've read, I like.
qcML is described in this paper from Waltzer et. al., and is currently in press (and open access) at MCP here.
I'd describe it, but I'm in a rush and this stolen image describes it pretty well!
Lets all do quality control! Not so much that we don't get experiments done, but enough that we know that our data is always good.
Sunday, April 27, 2014
100,000 views?!?!! What?
Short entry, as I'm pretty jetlagged. But while I was on vacation, the blog hit 100,000 views! Holy cow. Thanks for reading, everybody. And thanks for all the great comments that have been and keep rolling in!
Friday, April 25, 2014
Need another easy high yield digestion method? Try sTRAP!
Recently, I've heard of a number of a few groups who have had trouble with getting good peptide coverage using the FASP method. I'm still a big fan, but I realize that we might be trading reproducibility a little for sample digestion efficiency. There is obviously room, however, to improve on the FASP method and there are a lot of new variants out there.
In this month's Wiley Proteomics, Zougman et. al., go a completely different direction with their method, STrap. The method still seems relatively easy but maybe more robust than simple FASP. the article is open access (with Wiley registration) and worth checking out.
Thursday, April 24, 2014
Malaria proteomics -- no mass spec necessary
Cool new paper in EUPA Proteomics. This comes from Bachman et. al., and details the use of a protein array to study over 1,000 proteins in children with varying degrees of malaria. They end up finding a number of known and somewhat unknown (muscle proteins? weird) proteins that appeared differentially regulated with malaria severity.
An overview of the method employed is above.
Wednesday, April 23, 2014
SpliceVista. Find your sequence variants
More than 90% of Human genes have been found to have splice variants. Since we're still looking at the same old Uniprot, we miss most of them.
Enter SpliceVista from Yafeng Zhu et. al., from Karolinska (grab it while its still open access). Oh, and download it from GitHub here.
Whats it do? It grabs splice variant data that has been compiled by awesome new sequencing technologies and makes a new peptide database to compare your MS/MS spectra against?
Does it work? Sure looks like it! They process some acquired data and pull out almost a thousand MS/MS spectra that can be explained by splice variants, several of which are known to occur in these cell lines.
Lets figure out what all those unmatched MS/MS spectra are. They can't all be contaminants!
Tuesday, April 22, 2014
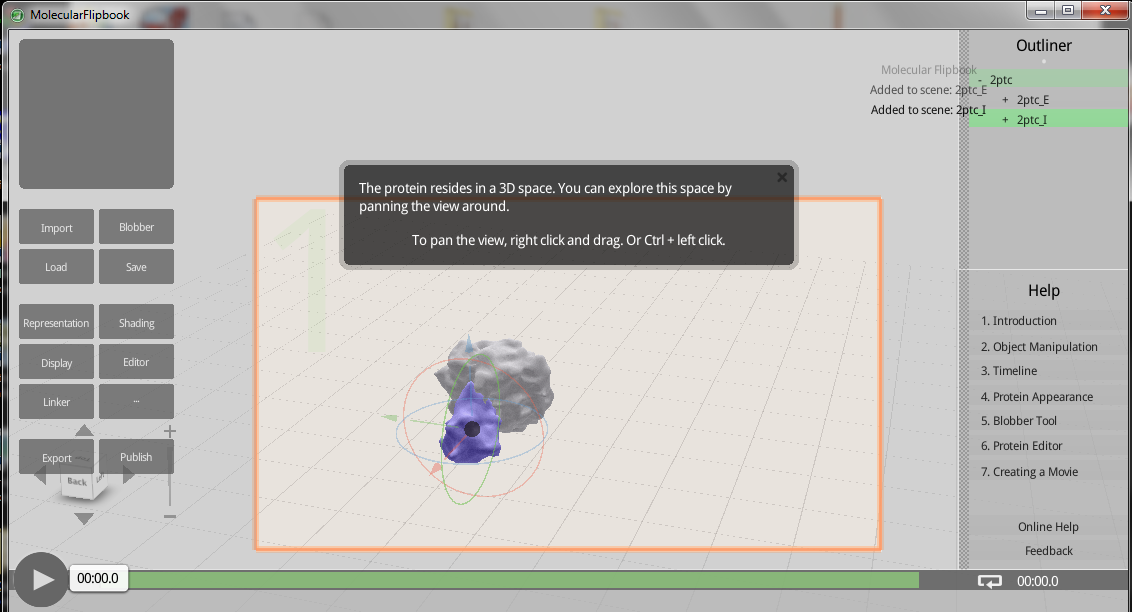
Molecular Flipbook! Free 3D protein modeling
Want to do 3D protein modeling? Just need a reason to buy cheesy 3D glasses from Amazon? Then I have a program for you! The Molecular Flipbook can do this for you. Directly upload proteins from the PDB.
You can read more about this software, as well as how to download it here.
Credit goes to Brenda for finding this one!
Thursday, April 17, 2014
Its official! My first ASMS!
Officially registered! After a decade or so in the field, I've given up. I'm a mass spectrometrist. There, I've said it. And as such, I should probably go to that thing y'all are always talking about. This year its almost literally in my back yard so I don't have much of an excuse.
See y'all in Baltimore, Hon!
Wednesday, April 16, 2014
wiSIMDIA -- the ultimate quantitative technique?
We all know the Fusion is fast. Crazy fast. And we know that the Fusion has some FTICR level resolution capabilities (though much faster). What if the crazies out at the Thermo factory decided to re-approach DIA (sometimes called SWOTH, or something) and utilized all of the powers of the Fusion to negate all of my complaints about these approaches?
Well, you'd probably end up with this technique described in this application note. I cut the core experiment description about above. Here is the gist: you do SIM at 240,000 resolution (1/4 million!) and break it into 3 largish windows. So you start with the depth and sensitivity of a SIM scan. At the same time, you have the ion trap do little DIA experiments of 12 Da windows, so you have the specificity of tiny DIAs on top of the dynamic range and specificity of big window SIMs.
If you bring me data where you show me your peptide of interest resolved at 240,000 resolution, 99% of the time I'm going to say "sure, that's your peptide" if you can back it up with 8 fragment ions from your DIA (which is the default quan method that this method uses!!!!!) from the same freaking experiment, you've identified that ion without any shadow of a doubt.
This method will be a default template in the next release of the Fusion software.
Update 4/16/14: There appears to be a problem with the graphics on the link above. The original PDF of the application note is available on Planet Orbitrap.
Tuesday, April 15, 2014
Virginia Tech still kicks ass!
When I chose Virginia Tech for graduate school I did it for 2 reasons, 1) Virginia Tech is a kick ass research school and is consistently one of the best science/engineering schools on the East Coast. (and 2) cause I love mountains! and it is one of the most beautiful locations I've ever seen in my life.
I was there seven years ago when a little prick took a gun and shot a bunch of nice people having class. As bad as that was, something that makes me almost equally angry is the fact that one day has almost overshadowed the fact that this is a premier research school. Prior to April 16th, 2007 Googling Virginia Tech would take you first to the school's rankings (and then to the football program). Since, it shows you first pictures of that little shit.
Off my soapbox, I'd like to use today's entry to remind people that Virginia Tech kicks ass and we need to forget about what-his-name.
As evidence, I'd like to point out that VT dumped $450M into research in 2011 (the newest numbers I could find). That places it cleanly into the top50 schools in this country. I'd also like to point out that, although it isn't super high on the NIH research institute rankings, tons of this money comes from private enterprises, but we also pull in the federal funding:
Dr. Chang Lu wont this NIBIB award this week to develop assays using as few as 100 cells to track disease progress.
Dr. Lu's grant will add to these numbers from last fall from the Carilion Research Institute (which didn't exist way back when I graduated).
SO. I'm getting off the soap box for real, and getting back to the science, but what I want to leave you with is this thought. The next time you think about Virginia Tech, think about a great school with a history and record of success in all sorts of research areas (and that appears more successful all the time) and not about something unfortunate that happened there. I don't want to ignore that event, but I don't want it to be all we consider when we hear the name of that awesome place.
Now all we need is some Orbitraps in there (don't worry, its one of my side projects.)
Monday, April 14, 2014
Assessment of MS/MS Search Algorithms with Parent-Protein Profiling
Okay, this is pretty smart! This paper is in this issue of JPR and from Miin Lin et al., at Wesleyan Connecticut.
There is inherently some uncertainty in the assignment of peptide/protein ID from shotgun proteomics data. What if we had a metric for it? In a decidedly old school and awesomely valid way of looking at it, these researchers went back to the tried and true molecular weight determinations from a nice old SDS-PAGE gel.
They cut slices so they knew the parent protein molecular weight, but dumped that data for now. They then used a slew of search algorithms with a 1% FDR and went back to see how well their peptide IDs corresponded to their molecular weights.
I think there is a possible criticism of this technique based on unknown cleavage products and post translational modifications causing shifts in the molecular weight of the protein or in the charge, and therefore shifting the pattern of protein migration. I would counter that argument by stating that I would expect this would be relatively minor in comparison to the high abundance proteins. I'm sure cleavage/PTMs have an effect but I don't think it hurts what an elegant analysis this is.
What were the conclusions? That using multiple algorithms give you a better shot of matching the expected protein identity. So, if you haven't been convinced already to use as many processing algorithms as practically possible to dig through your RAW data, here is yet another data point!
Sunday, April 13, 2014
Saturday, April 12, 2014
Deplete plasma? New info enters the fray.
To deplete or not to deplete -- part 2
In another of my favorite proteomics controversies, a new depletion technique takes a swing at whether depleting plasma is a good idea or not.
In this study, Seong-Beom Ahn and Alamgir Khan look at the impact depleting plasma has on cytokine recovery. Since cytokines are generally pretty low abundance, it makes for a decent metric. The depletion they use is an immuno-based one using magnetic beads.
Unlike other methods, this pull down approach showed either no impact or improved recovery of the cytokines. Definitely worth checking out!
This is what you get if you Google "pug hamlet"
Friday, April 11, 2014
Everything you ever wanted to know about any enzyme ever
Curious about a weird enzyme? Expasy has a page with a summary of the info on every proteolytic enzyme ever. Or at least a list with every one I've ever seen as well as a hundred more pages of info.
Check it out here.
Thursday, April 10, 2014
Monitor mutations via mass spec...in the clinic!
Monitoring mutations in clinical samples is becoming routine...using genetics approaches. What if we could do it by mass spectrometry? That would be a pretty big deal. Why? Because protein is what does stuff! Mutations in DNA may not actually result in protein production due to missense and nonsense mutations that end up in proteins that are cleaved, or not even expressed (respectively).
Sure, I'm biased, but I want to see real proteins that are messed up. And it is hard to do. Proteomics requires databases in most cases.
Thats why this paper from Dasari et. al., out of the Mayo Clinic is so awesome. Here we see a 2-pronged attack on amyloid (no, I didn't know what an amyloid deposit was until I looked it up) mutations coming out of a certified clinical lab.
How'd they do it?
Like this! They started with a super fasta including sequences from known mutations. The MS/MS spectra were searched with the super fasta vs 3 search engines. The MS/MS spectra were also searched with Tag Recon and all the data was compiled. Boom! Extensive coverage of known mutations along with novel mutations pulled out from the super cool Tag Recon package.
Yes, this seems super elaborate. It is. Roman Zenka is listed as an author, and I feel like you can clearly see his hand in this workflow.
Most of us don't have the programming/bioinformatics resources the Mayo Clinic does, but this is a shining example of how deep we can search our data when we really really want to.
Tuesday, April 8, 2014
Sweet new method for N-glyco peptide enrichment!
In press at MCP (currently, though not permanently, open access!) is this cool new paper from Ronghu Wu's lab at Georgia Tech.
In this study, the Wu lab ditches conventional enrichment techniques for a new 2-stage chemical approach: the glycos are reacted with boronic acid. The reaction is strong and reversible and the boronic derived glycos are easily enriched. Treating the enriched samples with PGNase F leaves samples that are easily analyzed by mass spec!
The end result? 816 N-glycosylation sites in yeast and 675 that were localized with greater than 99% confidences!
This is why we will always need chemists doing mass spec, regardless of how successful we are as biologists, sometimes we need chemists to go back and reinvent the techniques that have become so standard to us.
Sunday, April 6, 2014
Byonic + ptmRS
You probably saw this one coming. Last week I think I added a total of 7 new nodes to PD 1.4 on this laptop (details on the other 4 are coming, don't worry!). You know I'm biased, but I really believe that PD is rapidly becoming the most powerful and flexible proteomics package ever. Yes, this is heresy. I know the extreme love that a lot of you feel for the formerly great Elucidator. But when you see everything that PD can currently do (and will soon be able to, hint hint...) I think we'll have something that will blow even that legendary software package away.
Obviously, I've been obsessing a little over these two in specific: the Byonic node and ptmRS. How well do they work together? Are they redundant at all? Turns out they are perfectly and amazingly complementary!
A big part of my weekend has been Byonic wild card searching RAW files that my collaborator and I have acquired over the last 2 years in the search for PTMs that might explain our low peptide identification rates in certain files. (Yes, this is what I do for fun sometimes. I blast death metalall weekend, drink nice cheap Riojas and search RAW files with cool programs y'all send me. Yes, my new next door neighbors have already stopped talking to me...which is a shame cause they have a T.A.R.D.I.S bumper sticker and I figured we'd be friends...sometimes we just have to make sacrifices in the name of science!)
What I have found is a slight weakness in the Byonic node. I emphasize slight because I haven't actually read the instructions. I might be able to fix it by doing so. I have found that the Byonic node sometimes can't exactly assign a new PTM that it has found and you occasionally get multiple peptide options for the PTM locations. Easy example:
Byonic wild card search to find the PTMs that are present, followed by directly searching for those PTMs with a more conventional search engine:
Just another advantage of developers really grabbing on to the Discoverer frame work. We can not only get rid of apparent limitations in one algorithm, but also make strides toward turning it into an advantage!
Obviously, I've been obsessing a little over these two in specific: the Byonic node and ptmRS. How well do they work together? Are they redundant at all? Turns out they are perfectly and amazingly complementary!
A big part of my weekend has been Byonic wild card searching RAW files that my collaborator and I have acquired over the last 2 years in the search for PTMs that might explain our low peptide identification rates in certain files. (Yes, this is what I do for fun sometimes. I blast death metal
What I have found is a slight weakness in the Byonic node. I emphasize slight because I haven't actually read the instructions. I might be able to fix it by doing so. I have found that the Byonic node sometimes can't exactly assign a new PTM that it has found and you occasionally get multiple peptide options for the PTM locations. Easy example:
For this PSM, Byonic wild card search can't tell where the +57 mod is. Presumably, this is carbamidomethylation on the cysteine, so it should be the second one. The Byonic scores are virtually identical for the two options, so that isn't much help here. Easy to solve this one, but this could be more daunting if I, say a phosphorylation on site 223 in p53 means one thing and a phosphorylation on 229 means another....
Again, reading the instructions and/or changing some of the many settings we can control in Byonic may immediately improve this, but I have ptmRS!!!!!
Question 1) Are they compatible? As you can see from the screenshot at the top, Proteome Discoverer certainly believes that they are. When nodes aren't logical or compatible we can't draw arrows from one to the next. Starting a workflow results in no errors.
Unfortunately, however, the workflows do not finish. They end up crashing out. But thats okay. Because my laptop is FAST.
This is my current workflow of post translational modifications:
SequestHT (for high-low data) MSAmanda (for high high data) ran with ptmRS. While it would be useful to have the two programs running together, there is a big advantage for doing this way.
Confirmation! If Byonic tells you you are looking at a mod in your data and MSAmanda confirms that observation AND ptmRS gives you site localization that makes a ton of sense, how are you doing anything but winning?
Friday, April 4, 2014
Confetti. Everything you ever wanted to know about the HeLa proteome
As a field, we have a tendency to ignore what is going on in other fields. I understand, for real! This field is awesome. New technology, new software, new techniques. Its almost impossible to keep up with what is even happening in your favorite subsection of proteomics on your favorite PTM. However, there are times when we should probably lift our heads up and look around. Maybe that time is when people keep getting sued for studying and releasing data on a particular cell line.
So... its with a little hesitation that I write about this awesome compilation of data. The work is top notch. I wish it was on K562 or MCF-7 or one of the other amazing and well characterized cell lines out there. But it isn't. Its on HeLa, and despite my reservations, Confetti is a great contribution to our field and deserves to be acknowledged as such.
Off the soap box! This paper is currently open access at MCP, but won't be for long. So grab it here!
What they did: They extracted protein and digested it (with FASP! )in different combinations of 7 different enzymes. They used a variety of run techniques, including unfractionated and SAX fractionation to get a huge coverage. How huge?
8539 proteins.
44.7% coverage over the sequences of these 8539 proteins! Seriously. QExactive power!
Big deal? People have gotten numbers like this before. With UniprotHuman as the only database? Very very few people have ever obtained coverage this deep vs a manually annotated genome. Very few. Still not impressed?
Well, what if this group made a simple web application so that you could directly build and test SRMs off of this data set? Guess what. They did.
I'd put up a screenshot, but it is currently down. The paper hasn't officially been released yet, so the application may be going through some growing pains still. But the figures in the paper make it seem easy to use and powerful. Hopefully it will be up soon. Regardless, check out this paper. This is an idea of what we can get with proteomics when we really need to go deep and get awesome coverage.
When the application is back up, you can check it out here!
Update 4/15/14: The Confetti application had a minor web address issue, now resolved (see comments below). The resource is up and looks great!
Thursday, April 3, 2014
Multiplexed DIA on the Q Exactive
Okay, I've put this off long enough.
The final (so far!) method for quantification on the Q Exactive. Multiplexed DIA!
You can start this fun monologue on QE quan methods here.
This is what you get when the crazy smart people at the MacCoss lab take on new technology. Now, here is my disclaimer: I've never ran mutliplexed DIA (which I'll refer to as MSX-DIA) so I completely have to go from what I've been told and from the papers and presentations I've read (and brazenly stolen from).
Unlike the rest of this series, Skyline takes the top of the post over a photoshopped image of the Q Exactive. That's because you can't MSX-DIA without Skyline. You need it to set up the experiment and you need it to process the data.
I've already talked about DIA in part 4, so pick up this monologue from here if thats what you came here for.
The MacCoss lab took a step back from our simple DIA experiment and our 20 Da stepping windows (sometimes called swaths) and thought that the specificity could be improved. So they took the window and broke it into little tiny random chunks.
In this way, you can still cover the entire mass range because you've multiplexed random 4 Da windows together. Imagine that we've now taken my biggest objection to DIA (only knowledge of your peptide MS1 scan +/-20 Da and you ditched it. Thats what they did. Now, since we are multiplexing we are still getting fragment ions from loads of different precursors, but we've now randomized them.
Looking at the data directly doesn't do you a lot of good.
You need to de-multiplex it first. Whats that? How does it work? You've got me. Skyline can do it though. And if you doubt what Skyline can do, you probably haven't used it.
In the end, though, I know what you end up with. DIA without as much of the MS1 specificity argument. Since you are digging into 4Da windows (similar to a SIM) you get more dynamic range than you do out of your 20 Da windows. And from the data that has been published (in some pretty impressive journals) it appears to be reproducible and accepted. All good things!
And setting it up is easy (again, I haven't done it) but newer versions of Skyline have this nifty tab in it:
Multiplexed acquisition? Yep! And it outputs your "inclusion list" for the QE that provides your target windows.
And this is where I stop.
For more information, there is this sweet PPT from the MacCoss lab. (Warning: Direct download begins on clicking)
Thanks to Yue for explaining this to me and for the screenshots I stole from a talk she gave.
The final (so far!) method for quantification on the Q Exactive. Multiplexed DIA!
You can start this fun monologue on QE quan methods here.
This is what you get when the crazy smart people at the MacCoss lab take on new technology. Now, here is my disclaimer: I've never ran mutliplexed DIA (which I'll refer to as MSX-DIA) so I completely have to go from what I've been told and from the papers and presentations I've read (and brazenly stolen from).
Unlike the rest of this series, Skyline takes the top of the post over a photoshopped image of the Q Exactive. That's because you can't MSX-DIA without Skyline. You need it to set up the experiment and you need it to process the data.
I've already talked about DIA in part 4, so pick up this monologue from here if thats what you came here for.
The MacCoss lab took a step back from our simple DIA experiment and our 20 Da stepping windows (sometimes called swaths) and thought that the specificity could be improved. So they took the window and broke it into little tiny random chunks.
In this way, you can still cover the entire mass range because you've multiplexed random 4 Da windows together. Imagine that we've now taken my biggest objection to DIA (only knowledge of your peptide MS1 scan +/-20 Da and you ditched it. Thats what they did. Now, since we are multiplexing we are still getting fragment ions from loads of different precursors, but we've now randomized them.
Looking at the data directly doesn't do you a lot of good.
You need to de-multiplex it first. Whats that? How does it work? You've got me. Skyline can do it though. And if you doubt what Skyline can do, you probably haven't used it.
In the end, though, I know what you end up with. DIA without as much of the MS1 specificity argument. Since you are digging into 4Da windows (similar to a SIM) you get more dynamic range than you do out of your 20 Da windows. And from the data that has been published (in some pretty impressive journals) it appears to be reproducible and accepted. All good things!
And setting it up is easy (again, I haven't done it) but newer versions of Skyline have this nifty tab in it:
Multiplexed acquisition? Yep! And it outputs your "inclusion list" for the QE that provides your target windows.
And this is where I stop.
For more information, there is this sweet PPT from the MacCoss lab. (Warning: Direct download begins on clicking)
Thanks to Yue for explaining this to me and for the screenshots I stole from a talk she gave.
Wednesday, April 2, 2014
iORBI 2014: Full schedule!
For those of you in or near 20ish select US cities, its iORBI time! For those of you farther away, all of the information covered during this tour will be made fully available to you as well, albeit a bit later. You can always check out the information from iORBI 2012 in the meantime.
Never been to an iORBI? Unfamiliar? It is a meeting exclusively for Orbitrap experts. This isn't a sales or marketing meeting. Scientists inside Thermo provide uncensored talks on specific applications. When I say uncensored, I mean it. This is the "good the bad and the ugly," as it is sometimes said. How to optimize your applications, what downsides and drawbacks there are, how to address them, etc., Something that I've always found cool is the fact that the talks are tailored to specific cities. For example, Pittsburgh is starting to establish itself as the lipidomics capital of the U.S. so there is no way that we would skip having a great metabolomics expert talk about lipids there, where we might not give that talk in Ann Arbor. Don't worry lipid experts in Michigan, you'll have full access to the lipidomics talk with copious speaker notes, extra slides, and contact information on the experts who put the slides together.
Here is the full schedule (partially so its easy for me to find. You wouldn't believe the clutter on my desktop right now...). Not registered? Contact your local sales rep.
Madison, WI
|
T, April 8th
|
Rochester, MN
|
W, April 9th
|
Minneapolis, MN
|
Th, April 10th
|
Philadelphia, PE
|
T, April 22nd
|
Gaithersburg, MD
|
W, April 23rd
|
New Jersey/NY at Rutgers
|
Th, April 24th
|
Boston/Cambridge
|
W, April 30th
|
Ithaca, NY
|
Th, May 1st
|
Seattle, WA
|
M, May 5th
|
Genentech, CA
|
W, May 6th
|
Stanford, CA
|
T, May 7th
|
Los Angeles, CA
|
Th, May 8th
|
Indianapolis, IN
|
T, June 24th
|
Chicago, IL
|
W, June 25th
|
St. Louis, MO
|
Th, June 26th
|
Montreal
|
T, Sept 9th
|
Toronto
|
Th, Sept 11th
|
RTP, NC
|
T, Sept 16th
|
Pittsburgh, PA
|
T, Oct 21st
|
Ann Arbor, MI
|
W, Oct 22nd
|
Columbus, OH
|
Th, Oct 23rd
|
Austin, TX
|
T, Nov 11th
|
Houston, TX
|
Th, Nov 13th
|
Tuesday, April 1, 2014
Distraction: Overly honest methods
This guy at Imgur slapped together a his favorite entries from #overlyhonestmethods onto random lab images. The result is pretty hilarious.
You can find them at Imgur here.
Credit goes to Alexis for this one!
Subscribe to:
Comments (Atom)