I've often wondered who is using which search engines. I've also made guesses and they don't seem to be anywhere near accurate based on this Google Scholar analysis by Dr. Felipe da Veiga Leprevost who I finally met in person at US HUPO this year.
He shared this on social media and was clear that it wasn't comprehensive (who can keep up with the 1,000 plus proteomics tools out there? I mean, @PastelBio is doing an incredible job, but I don't think it's at 1,000 yet.)
This is by citations, so there are lags, and it doesn't tell us what companies that don't publish are doing (and there are a lot of instruments in those places). Click to expand.
Check out the overlay, though!
From this it looks like MaxQuant is, and has been, massively dominant for proteomics data analysis in the literature. The fastest growing engine out there, probably surprising few of us, is MSFragger. I didn't use FragPipe for the first time until 2019 and I don't think it had been out for very long when I started using it.
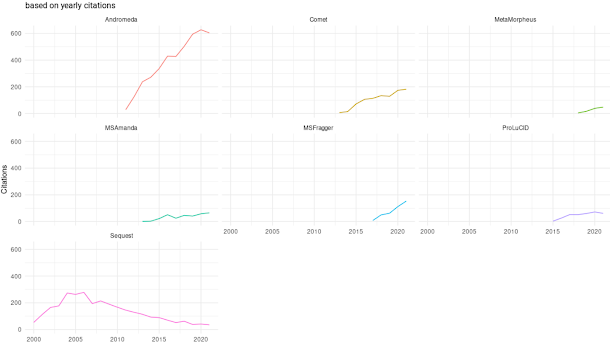

That is all well. But I wonder if the use of this different search engines and preference to one versus others (Andromeda vs Sequest in particular) is due to the fact that one is free and the other is commercially available at a high cost.
ReplyDeleteOver the years I have used different search engines, and I have found only slight differences among all of them. I might be wrong since I am not routinely testing every new improvement of all of them.
Is there then a criteria based not on popularity but on accuracy?