Punchline? Sometimes people who take HIV drugs have negative neurological effects like dementia. We don't know why. What Dr. Wheeler found was that if she took purified endoplasmic reticulum from different brain regions and treated them with HIV drugs, some metabolism occurred. Metabolism is supposed to happen in the liver, not the brain, right?
While class I metabolism (P450 type oxidation) had been previously observed, class II metabolism stuff really had not been, and that was what she was seeing. And that didn't make sense.
The QE couldn't detect enzymes capable of this process. DDA/DIA/PRM
The TIMSTOFs didn't detect enzymes capable of these processes (PASEF/diaPASEF).
Targeting on a TIMSTOF is a miserable experience. The files are huuuuuuuuuuuuge.
Enter ZenoTOF and ZenoPulsing enabled PRM, which they call MRMHR. MRMHR produces tiny output files that are super simple to process in Skyline with ridiculously high sensitivity.
The first author was able to demonstrate that enzymes matching the metabolites observed in the correct brain ER cell types all lined up. I can't remember if this made the paper, but mRNA could back it up as well.
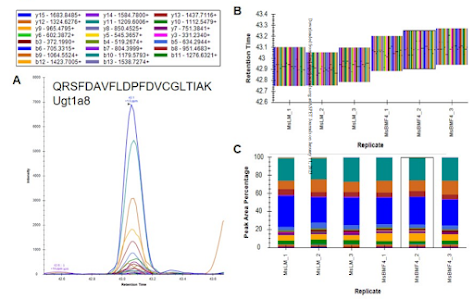
I know this didn't make the paper, but she also fractionated a bunch of peptides from these brain regions and did really comprehensive proteomics using the same instrument. Using the ProteomicRuler we were able to get some ballpark estimates for how rare the enzymes she is seeing are. Current best numbers are these are at less than 300 copies per cell. Which ain't much. Files are all on MASSIVE at MSV000090576.

No comments:
Post a Comment